|
Size: 21398
Comment:
|
Size: 30844
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
| This tutorial contains more advanced topics, such as focused refinement and classification, symmetry breaking, and analysis of continuous motion. For new users, it is recommended to start with a simple [[https://blake.bcm.edu/emanwiki/EMAN2/e2TomoSmall | tutorial]]. Many of the functions here require an EMAN2 installation after 04/2023. A latest continuous build may work but compile from source is preferred. For the installation guide, see [[https://blake.bcm.edu/emanwiki/EMAN2/Install/SourceInstall | here]]. |
This tutorial contains advanced topics of the SPT workflow, such as focused refinement and classification, symmetry breaking, and analysis of continuous motion. For new users, it is recommended to start with a simple [[https://blake.bcm.edu/emanwiki/EMAN2/e2TomoSmall | tutorial]]. Many of the functions here require an EMAN2 installation after 05/2023. A newer version is almost always better. A latest continuous build may work but compiling from source is preferred. For the installation guide, see [[https://blake.bcm.edu/emanwiki/EMAN2/Install/SourceInstall | here]]. |
| Line 15: | Line 15: |
| Make sure any EMAN2 commands you run are executed from within this folder (not any subfolder). | Make sure all EMAN2 commands you run are executed from within this folder (not any subfolder). |
| Line 19: | Line 19: |
| The files provided are already imported to EMAN2, so simply unzipping them in the project folder should work. For other CryoET projects, it is recommended to import the tilt series through e2projectmanager. Import tilt series will also convert them to hdf format, which greatly reduces storage (from 3.1GB to 700MB per tilt series) without obvious side effects. | The files provided are already imported to EMAN2, so simply unzipping them in the project folder should work. For other CryoET projects, it is recommended to import the tilt series through '''e2projectmanager'''. Import tilt series will also convert them to hdf format, which greatly reduces storage (from 3.1GB to 700MB per tilt series) without obvious side effects. After unzipping the files, you should have a folder called "tiltseries", with four hdf image stacks in it, inside the project folder. To view the tilt series, run '''e2display.py''', locate the file in the browser, and click '''Show2D'''. |
| Line 23: | Line 25: |
| Line 26: | Line 27: |
| Start from one good tilt series to play with the parameters for tilt series alignment and tomogram reconstruction. | Start from one good tilt series to play with the parameters for tilt series alignment and tomogram reconstruction. You often need to run this multiple times with different options to get a satisfying reconstruction. After having a good set of options, we will run the program with the same parameters for all tilt series later. |
| Line 32: | Line 33: |
| Always check --autoclipxy for non-square images. --patchtrack=2 now works almost as well as landmark based tracking when the fiducials are not present. It should not stop you from getting to ~3Å with enough particles and good samples. It also provides a good starting point for landmark based tracking so it does not hurt to include it. | * Always check --autoclipxy for non-square micrographs. The program will decide how to clip based on the content of the tomogram. * Patchtracking (--patchtrack=2) now works almost as well as landmark tracking (controlled by --niter=X) when the fiducials are not present. It should not stop you from getting to ~3Å with enough particles and good samples, even with --niter=0. Having both normally does not hurt. * Tomogram thickness needs to be decided visually. The default often works, but it is good to increase the number if there are still features at the top and bottom of the tomogram. |
| Line 39: | Line 42: |
| Before reconstructing all tilt series, we need to make sure the handedness of the tomogram is correct. |
|
| Line 45: | Line 50: |
| Temporary files are written in '''tomoctf_xx''' if `--writetmp` is specified. At high tilt, i.e. tilt ID close to 0 or the number of tilt, the curve of one of the handedness (blue) should show a much deeper dip than the other one (red). If it is not obvious, something in the earlier steps might be wrong and the images might not have enough signal for CTF estimation. Also, if the dip is too close to the left or right end of the plot, consider adjusting the range of defocus search using `--dfrange`. Note this only accounts for the geometry of the tilt series, but it can still produce the wrong handedness if your individual micrographs are flipped. This can sometimes be the case with some data collection software. Even in those cases, you should still use the handedness recommended by the program (or go back and flip the raw micrographs), which will produce more stable defocus estimation. |
The program will print out the recommended tilt axis angle from the command line, and we will reconstruct all tomograms using the new tilt axis angle (--tltax option in e2tomogram). The result relies on the fitting of CTF rings in the micrographs, and can be unreliable if there is not enough signal. Only believe the result when >80% of the tilt angles have the same handedness. It is better to also check the temporary files, which are written in '''tomoctf_xx''' if `--writetmp` is specified. At high tilt, i.e. tilt ID close to 0 or the 35, the curve of one of the handedness (blue) should show a much deeper dip than the other one (red). If it is not obvious, something in the earlier steps might be wrong and the images might not have enough signal for CTF estimation. Also, if the dip is too close to the left or right end of the plot, consider adjusting the range of defocus search using `--dfrange`. Note this only accounts for the geometry of the tilt series, but it can still produce the wrong handedness if your individual micrographs are flipped. This can sometimes be the case with some data collection software. Even in those cases, you should still use the handedness recommended by the program (or go back and flip the raw micrographs), which will produce a more accurate defocus estimation. |
| Line 58: | Line 63: |
| Now we estimate the defocus for all tilt series. Note here we only estimate the CTF, not correct for it. The correction will happen later when extracting particles. To get to high resolution, it is often safer to use a higher than default `--tilesize`. | Now we estimate the defocus for all tilt series. Note here we only estimate the CTF, not correct for it. The correction will happen later when extracting particles. To get to high resolution, it is often safer to use a higher than the default `--tilesize`. |
| Line 77: | Line 82: |
In this tutorial, we select particles using the automatic particle picker. This program is deep neural network based, and requires GPU and CUDA installed. Without CUDA, it is also possible to manually pick a few particles, generate initial model, then use that model for template matching. It should work almost as well since the virus is an easy target. Here we run |
|
| Line 80: | Line 87: |
| Since the virus particles are quite large, here we change the reference box size to 192 using the '''!ChangeBx''' button. Start from 5 positive and 5 negative references, train and apply the network, then refine the network by adding more false positives/negatives from multiple tomograms to the training set. Apply the trained network to all tomograms then close the window. Here we get 300 particles from 4 tomograms. | Since the particles are quite large, we change the reference box size to 192 using the '''!ChangeBx''' button first. Start from 5 positive and 5 negative references, train and apply the network, then refine the network by adding more false positives/negatives from multiple tomograms to the training set. Apply the trained network to all tomograms then close the window. Here we get ~300 particles from 4 tomograms. |
| Line 84: | Line 92: |
| After auto-boxing, we can take a look at the particles using '''e2spt_boxer.py''' to make sure they are correct, and also decide the correct box size. | |
| Line 87: | Line 96: |
| Extract particles of the P22 capsids. For speed and memory consumption, we use a box size of 640 and shrink the particles by 4 using `--shrink`. {{{ e2spt_extract.py --boxsz_unbin=640 --label=p22 --threads=32 --alltomograms --shrink=4.0 --tltkeep=1.0 --compressbits=8 --parallel thread:8 |
After selecting all particles, we extract particles of the P22 capsids. For speed and memory consumption, we use a box size of 640 and shrink the particles by 4 using `--shrink`. After extraction, run '''e2spt_buildstack.py''' to generate a virtual stack of all particles of the same time from different tomograms. {{{ e2spt_extract.py --boxsz_unbin=640 --label=p22 --threads=8 --alltomograms --shrink=4.0 --tltkeep=1.0 --compressbits=8 --parallel thread:4 |
| Line 94: | Line 103: |
| == Refinement workflow == {{attachment:workflow.gif|Refinement workflow|width="600"}} |
|
| Line 96: | Line 109: |
| Since we start from the icosahedral capsid which does not have many low resolution features, here we set `--sym=icos` and `--res=30`. Leave the rest of the options as default. {{{ e2spt_sgd_new.py sets/p22_bin4.lst --res=30.0 --niter=100 --shrink=1 --parallel=thread:17 --ncls=1 --batch=12 --learnrate=0.2 --sym=icos |
We start by generating the initial model directly from the particles. Since the icosahedral capsid does not have many low-resolution features, here we set `--sym=icos` and `--res=20` to target a higher resolution. {{{ e2spt_sgd_new.py sets/p22_bin4.lst --res=20.0 --niter=100 --shrink=1 --parallel=thread:17 --ncls=1 --batch=12 --learnrate=0.2 --sym=icos |
| Line 103: | Line 116: |
| Depending on the quality of particles, there can be some randomness in the initial model generation process, and the command can sometimes converge to a featureless sphere. A safer option is to set `--ncls=3`, so the program will generate 3 initial models at the same time, but this will also be slower. |
|
| Line 105: | Line 120: |
| Start from the symmetrical refinement of the capsid. This should reach Nyquist of the particles (17Å) easily. One thing to note here is the FSC curve should improve after the subtilt refinement (red vs blue curve below). If it does not, it means either the data does not have enough signal per particle per tilt for the alignment, which may greatly limit the resolution achieved, or the tilt series alignment is so good that there isn’t much to improve, which does not really happen often. | Start from the symmetrical refinement of the capsid. Here we use the "new" refinement routine in '''e2spt_refine_new.py''' that integrates the subtomogram (`p` iterations) and subtilt refinement (`t` and `r` iterations). The refinement should reach Nyquist of the particles (17Å) easily. One thing to note here is the FSC curve should improve after the subtilt refinement (red vs blue curve below). If it does not, it means either the data does not have enough signal per particle per tilt for the alignment, which may greatly limit the resolution achieved, or the tilt series alignment is so good that there isn’t much to improve, which does not really happen often. |
| Line 113: | Line 128: |
| == Finding tail using symmetry breaking == After the icosahedral capsid refinement, we have two main goals here. One is to push the symmetrical capsid structure toward a higher resolution, and the other is to find the tail of the virus and solve its structure. However, the two goals are somewhat intertwined, and we will need to get back and forth a bit to solve both. Here we first look for the tail of the phage with symmetry breaking. Start by running '''e2spt_refinemulti_new.py''' targeting a low resolution (--maxres=80). {{{ e2spt_refinemulti_new.py --nref 1 --ptcls spt_00/aliptcls3d_06.lst --niter 5 --maxres 80 --breaksym icos --loadali3d --parallel thread:32 --sym c5 --skipali --loadali2d spt_00/aliptcls2d_08.lst }}} Here we load both the 3D subtomogram orientation assignment and subtilt alignment from the previous refinement folder. Since we break the icosahedral symmetry and re-apply the c5 symmetry, the densities of the tail should show up the penton at either the top or the bottom of the capsid after a few iterations. One easy way to visualize this is to look at the central slice views. In the e2display browser, select the map and click '''RngXYZ'''. Here we set '''Sum layers''' to 10 to look at a 20-pixel thick slice. {{attachment:symbreak0.png|Symmetry breaking 1|width="600"}} Now we have an initial capsid structure with a tail, we can do another run of the symmetry breaking using this as the reference, and target a higher resolution. To ensure convergence, we would like to craft a mask that covers only the location of the tail we have in the reference. It is more convenient to make the mask in Filtertool, which can be launched by selecting the reference map ('''sptcls_00/threed_05_00.hdf'''), and clicking '''Filtertool''' in the '''e2display''' browser. If you have a slow machine and the program crashes often, hold Shift and click the button to enter safe mode. In the Fitertool, first add a soft mask using '''mask.soft''', and change '''outer_radius''' and '''dz''' so it covers only the density of the tail. Afterward, add a '''math.linear''' processor before the soft mask, which will set the map to all ones, so the output would be a spherical mask with a soft edge instead of masked-out density. Save the mask using '''File->Save''', then rename it to '''mask_tail0.hdf'''. Finally, we add the mask of the capsid to the mask of the tail to make sure the alignment does not drift away from the initial capsid refinement. {{attachment:tailmask0.png|mask tail 0|width="600"}} {{{ e2proc3d.py mask_tail0.hdf mask_tail1.hdf --addfile spt_00/mask.hdf --process threshold.clampminmax:maxval=1 }}} Here we also add a '''threshold.clampminmax''' processor to make sure there is no value above 1 in the mask. {{attachment:symbreak2_msk.png|Symmetry breaking msk|width="600"}} With the mask ready, we can now start the second symmetry breaking run. The program will be slower since we took out the `--skipali` option, and will do local orientation refinement in addition to symmetry breaking. {{{ e2spt_refinemulti_new.py sptcls_00/threed_05_00.hdf --ptcls spt_00/aliptcls3d_06.lst --niter 10 --maxres 30 --breaksym icos --loadali3d --parallel thread:32 --minres 150 --sym c5 --maskali mask_tail1.hdf --loadali2d spt_00/aliptcls2d_08.lst }}} {{attachment:symbreak2.png|Symmetry breaking 2|width="600"}} Since we are applying the c5 symmetry here, the tail will not be fully resolved, but the location of the tail should be mostly correct, which is indicated by the solid density in the structure after the refinement. Now that we found the tail of each capsid, we can re-extract the unbinned particles of the tail base on the refinement results. In particle extraction, ignore the tomogram options, and only input the '''aliptcles3d_xx.lst''' through the `--jsonali` option. The program will find the particles given the refinement results. The program also allows extracting particles with an offset from the previous refinement, so here we can specify `--postxf=icos,0,0,-50`, where (0,0,-50) is the coordinates of the tail from the averaged structure. At this point, we will only extract the particles and build sets (this will give us '''sets/tail.lst'''), but do not refine the tail structure yet. {{{ e2spt_extract.py --boxsz_unbin=208 --newlabel=tail --threads=8 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=sptcls_01/aliptcls3d_10_00.lst --compressbits=8 --postxf c1,0,0,-50 --mindist 100 --parallel thread:4 e2spt_buildsets.py --allparticles --label tail }}} |
|
| Line 115: | Line 173: |
| Now we extract particles at each penton of the capsid to refine them locally. In particle extraction, ignore the tomogram options, and only input the '''spt_xx/aliptcles3d_xx.lst''' through the `--jsonali` option. The program will find the particles given the refinement results. Here we want to get all 12 pentons for each capsid, so we specify --postxf=icos,0,0,35, where (0,0,35) is the coordinates of one penton from the capsid refinement. {{{ e2spt_extract.py --boxsz_unbin=190 --newlabel=capc5 --threads=32 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=spt_00/aliptcls3d_03.lst --compressbits=8 --postxf icos,0,0,35 --mindist 100 --parallel thread:32 e2spt_buildsets.py --allparticles --label capc5 }}} After extraction, instead of starting a refinement directly, gather the metadata of the newly extracted particles using the script below. The program will collect the information from particle headers so the new particles inherit the orientation and even/odd split of the previous refinement. With the `--ali2d` input, it also interpolate the subtilt movement of each new 2D particle from the movement of particles in the same tilt from the previous alignment. {{{ e2spt_gathermeta.py --ptcls sets/capc5.lst --ali2d spt_00/aliptcls2d_04.lst |
Now we get back to the icosahedral capsids and extract particles at each penton of the capsid to refine them locally. The particle extraction program can generate multiple new particles from each original one with a given symmetry. Here we want to get all 12 pentons for each capsid, so we specify `--postxf=icos,0,0,35`, where (0,0,35) is the coordinates of one penton from the capsid refinement. Make sure to also set a number for `--mindist`, which defines the minimum distance between the new particles. Here the program will initially find 60 new penton particles for each capsid particle based on the icosahedral symmetry, then reduce the number to 12 since every 5 particles would have the same coordinates. {{{ e2spt_extract.py --boxsz_unbin=192 --newlabel=pent --threads=8 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=spt_00/aliptcls3d_03.lst --compressbits=8 --postxf icos,0,0,35 --mindist 100 --parallel thread:4 e2spt_buildsets.py --allparticles --label pent }}} After extraction, instead of starting a refinement directly, we first gather the metadata of the newly extracted particles using the '''e2spt_gathermeta.py''' program. The program will collect the information from particle headers so the new particles inherit the orientation and even/odd split of the previous refinement. With the `--ali2d` input, it also interpolates the subtilt movement of each new 2D particle from the movement of particles in the same tilt from the previous alignment. Also note that out of the 12 pentons in each capsid, the penton with the tail would have a different structure than the others. Out of an abundance of caution, we do not use all pentons we just extracted, but remove the ones that are close to the tails we found earlier. This is done by providing the tail particles through the `--exclude` option, which is why we need to find the tails before this step. In practice, since it is only 1/12 of the entire population, the pentons with tails will likely be excluded or down-weighted in the iterative refinement, and won't actually cause any difference. But it is still a safer practice to avoid potential issues down the way. {{{ e2spt_gathermeta.py --ptcls sets/pent.lst --ali2d spt_00/aliptcls2d_05.lst --exclude sets/tail.lst |
| Line 140: | Line 200: |
| == Sharpening using structure factor == As we are achieving subnanometer resolution, it is often helpful to sharpen the map to get better real space features. Here we can run {{{ e2spt_structfac.py --even spt_01/threed_raw_even.hdf --res 7 }}} This will fit B-factors based on the given map and the classical protein power spectrum, and create a 1D curve (sf.txt by default) that can be used to sharpen the structure. To visualize the results, copy the unfiltered maps to another iteration number, and rerun the last e2refine_postprocess command with the text file. {{{ cp spt_01/threed_raw_even.hdf spt_01/threed_10_even.hdf cp spt_01/threed_raw_odd.hdf spt_01/threed_10_odd.hdf e2refine_postprocess.py --even spt_01/threed_10_even.hdf --setsf sf.txt --tophat localwiener --threads 32 --restarget 7 --sym c5 }}} Once created, the file can be input to other e2spt_refine_new.py runs through --setsf option to sharpen the map at every iteration. |
|
| Line 161: | Line 202: |
| Run the following command to evaluate the results of a subtilt refinement. | After the refinement (or at least one `t` iteration) is finished, we can run the following command to evaluate the results of a subtilt refinement. |
| Line 168: | Line 209: |
| Typically, you should see the amount of movement in each tilt (red dashed line) roughly correlated with the particle score (blue line), both of which increase at higher tilt. In each tilt, the vectors should form a somewhat locally correlated flow field. Note the length of arrows are exaggerated, since they normally do not move at a length that’s visible at the scale of the entire tomogram. == Finding tail using symmetry breaking == Now get back to the capsid refinement and look for the tail of the phage with symmetry breaking. Start by making a mask covering one of the pentons along the c5 axis. It is more convenient to make the mask in Filtertool, simply using a '''math.linear''' processor to set the map to all ones, then setting the mask using '''mask.soft'''. Save the mask using '''File->Save''', then rename it to '''mask_pent0.hdf'''. {{attachment:make_mask.png|Make mask|width="600"}} While running symmetry breaking with just this mask may work, it can be a bit tricky since the program can actually put any penton at the location of the mask without a given reference. One way to get around this is to use a mask that covers two pentons at the opposite sites, and break the icosahedral symmetry while applying the c5 symmetry, so the program will put the two pentons with the most differences at the two sites, and one of them will have the tail. So run {{{ e2proc3d.py mask_pent0.hdf mask_pent1.hdf --rot 0,180,0 e2proc3d.py mask_pent0.hdf mask_pent2.hdf --addfile mask_pent1.hdf }}} Then, run {{{ e2spt_refinemulti_new.py --nref 1 --ptcls spt_00/aliptcls3d_03.lst --niter 10 --maxres 40 --breaksym icos --loadali3d --parallel thread:32 --minres 200 --sym c5 --maskali mask_pent2.hdf --skipali }}} In a few iterations, vague densities of the tail should show up at one side of the capsid. One easy way to visualize this is to look at the central slice views. In the e2display browser, select the map and click '''RngXYZ'''. Here we set '''Sum layers''' to 10 to look at a 20 pixel thick slice. {{attachment:symbreak1.png|Symmetry breaking 1|width="600"}} Then we can do another run of the symmetry breaking using this as the reference, and the corresponding mask that covers the tail. Note here we do not add the `--skipali` option so the program will do some local alignment at the tail. To make sure the alignment does not drift too far away, we include the capsid into the mask as well. {{{ e2proc3d.py mask_pent0.hdf mask_pent0x.hdf --addfile spt_00/mask.hdf --process threshold.clampminmax:maxval=1 }}} There is about a 50/50 chance that the tail density appears at the top or bottom, so pick either mask_pent0.hdf or mask_pent1.hdf based on the result. After adding the capsid mask from '''spt_00''', use '''threshold.clampminmax''' to make sure there is no value above 1 in the mask. {{attachment:symbreak2_msk.png|Symmetry breaking msk|width="600"}} {{{ e2spt_refinemulti_new.py sptcls_00/threed_05_00.hdf --ptcls spt_00/aliptcls3d_03.lst --niter 10 --maxres 40 --breaksym icos --loadali3d --parallel thread:32 --minres 200 --sym c5 --maskali mask_pent0x.hdf }}} {{attachment:symbreak2.png|Symmetry breaking 2|width="600"}} Since we are applying the c5 symmetry here, the tail will not be fully resolved, but the location of the tail should be mostly correct, which is indicated by the solid density in the structure after the refinement. |
Typically, you should see the amount of movement in each tilt (red dashed line) roughly correlated with the particle score (blue line), both of which increase at higher tilt. In each tilt, the vectors should form a somewhat locally correlated flow field. Note the length of arrows is exaggerated, since they normally do not move at a length that’s visible at the scale of the entire tomogram. |
| Line 213: | Line 213: |
| Now we found the tail of each capsid, we can re-extract the particles of the base on the refinement results. To solve the structure, we still need to break the c5 symmetry of the capsid and apply the c6 symmetry of the tail. {{{ e2spt_extract.py --boxsz_unbin=200 --newlabel=tail1 --threads=32 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=sptcls_01/aliptcls3d_10_00.lst --compressbits=8 --postxf c1,0,0,50 --mindist 100 --parallel thread:32 e2spt_buildsets.py --allparticles --label tail1 e2spt_refinemulti_new.py --nref 1 --ptcls sets/tail1.lst --niter 10 --maxres 40 --breaksym c5 --loadali3d --parallel thread:32 --minres 200 --sym c6 --skipali |
Now back to the tail refinement. Since we now have the high resolution of the capsid pentons, we will use the subtilt alignment information from there to guide the refinement of the tail. But first, we still need to break the c5 symmetry we imposed previously and re-apply the c6 symmetry, to get an initial structure of the tail. {{{ e2spt_refinemulti_new.py --nref 1 --ptcls sets/tail.lst --niter 10 --maxres 30 --breaksym c5 --loadali3d --parallel thread:32 --minres 200 --sym c6 --skipali |
| Line 225: | Line 221: |
| Then, make a mask for the tail using Filtertool, call it '''mask_tail1.hdf'''. | Before refining the tail, we would like to make a customized mask for it, instead of using the auto masking routine in EMAN2. The goal here is to remove the impact of the capsid density which has c5 symmetry to prevent it from impacting the alignment. Again, we make this using the Filtertool. Here we also start from a '''mask.soft''' processor to get to rough location of the tail, then add a '''mask.auto3d.thresh''' processor after it to generate the mask. This processor selects all voxels at threshold2 that are connected to the voxels above threshold1, and adds extra padding afterward. We rename this mask '''mask_tail1.hdf'''. |
| Line 233: | Line 229: |
| e2spt_gathermeta.py --ptcls sets/tail1.lst --ali2d spt_01/aliptcls2d_07.lst --ali3d sptcls_02/aliptcls3d_10_00.lst | e2spt_gathermeta.py --ptcls sptcls_02/aliptcls3d_10_00.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d |
| Line 239: | Line 235: |
| e2spt_refine_new.py --path spt_02 --continuefrom 0.5 --sym c6 --breaksym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --setsf sf.txt --iters=p4,t2 --keep=0.95 --threads=32 --mask mask_tail1.hdf }}} {{attachment:tail_refine1.png|Tail refinement 1|width="600"}} |
e2spt_refine_new.py --path spt_02 --continuefrom 0.5 --sym c6 --breaksym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --iters=p3,t2,p,t --keep=0.95 --threads=32 --mask mask_tail2.hdf }}} {{attachment:tail_refine1.png|Tail refinement 1|width="300"}} |
| Line 246: | Line 242: |
| The single model refinement will get to ~20Å resolution. One limitation of the refinement is the heterogeneity at the lower part of the tail where it interacts with the membrane. This is probably because only some of the particles are anchored on the membrane, whereas the others are floating in solution. To sort them out, do some more classification. Similarly, we create a soft mask that covers the lower part of the tail, and call it mask_tail0.hdf {{{ e2spt_refinemulti_new.py --nref 2 --ptcls spt_02/aliptcls3d_05.lst --niter 10 --maxres 50 --loadali3d --parallel thread:32 --minres 500 --sym c6 --maskali mask_tail0.hdf --skipali |
The single model refinement will get to ~15Å resolution. One limitation of the refinement is the heterogeneity at the lower part of the tail where it interacts with the membrane. This is likely because only some of the particles are anchored on the membrane, whereas the others are floating in the solution. To sort them out, do some more classification. Similarly, we create a soft mask that covers the lower part of the tail, and call it mask_tail3.hdf {{{ e2spt_refinemulti_new.py --nref 2 --ptcls spt_02/aliptcls3d_07.lst --niter 10 --maxres 25 --loadali3d --parallel thread:32 --minres 200 --sym c6 --maskali mask_tail3.hdf --loadali2d spt_02/aliptcls2d_08.lst --skipali |
| Line 256: | Line 252: |
| It is also possible to further refine the structure of individual classes. {{{ e2spt_gathermeta.py --ptcls sptcls_03/aliptcls3d_10_01.lst --ali2d spt_03/aliptcls2d_07.lst --ali3d sptcls_03/aliptcls3d_10_00.lst e2spt_refine_new.py --path spt_03 --continuefrom 0.5 --sym c1 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --setsf sf.txt --iters=p4,t2,p,t,r --keep=0.95 --threads=32 --mask mask_tail1.hdf }}} {{attachment:tail_refine2.png|Tail refinement 2|width="600"}} This will produce a map at ~16Å resolution, slightly better than the previous refinement despite having ~30% fewer particles. The channel through the membrane becomes more visible too. We can still run some further classification from here, focusing on the protein-membrane interacting region and targeting a slightly higher resolution. {{{ e2spt_refinemulti_new.py --nref 2 --ptcls spt_05/aliptcls3d_05.lst --niter 10 --maxres 30 --loadali3d --parallel thread:32 --minres 200 --sym c6 --maskali mask_tail2.hdf }}} {{attachment:tail_classify2.png|Tail classification 1|width="600"}} Here we can see a movement that looks like the opening and closing of the channel, which seems to correlate with the outward tilting of the tail fibers. |
It is also possible to further refine the structure of individual classes. This will give us better features at the membrane channel, but the resolution will be limited by particle count. {{{ e2spt_gathermeta.py --ptcls sptcls_03/aliptcls3d_10_01.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d e2spt_refine_new.py --path spt_03 --continuefrom 0.5 --sym c6 --breaksym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --iters=p2,t2,p,t --keep=0.95 --threads=32 --mask mask_tail2.hdf }}} {{attachment:tail_refine2.png|Tail refinement 2|width="300"}} |
| Line 286: | Line 269: |
| cp spt_05/aliptcls3d_05.lst spt_05/aliptcls3d_05_c6.lst e2proclst.py spt_05/aliptcls3d_05_c6.lst --sym c6 |
e2proclst spt_03/aliptcls3d_05.lst --create spt_03/aliptcls3d_05_c6.lst e2proclst.py spt_03/aliptcls3d_05_c6.lst --sym c6 |
| Line 293: | Line 276: |
| e2spt_gathermeta.py --ptcls spt_05/aliptcls3d_05_c6.lst --ali2d spt_03/aliptcls2d_07.lst --ali3d spt_05/aliptcls3d_05_c6.lst e2spt_refine_new.py --path spt_09 --continuefrom 0.5 --sym c1 --parallel thread:32 --startres 30 --localrefine --setsf sf.txt --iters=p3 --keep=0.95 --threads=32 --mask mask_tail5.hdf |
e2spt_gathermeta.py --ptcls spt_03/aliptcls3d_03_c6.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d e2spt_refine_new.py --path spt_04 --continuefrom 0.5 --sym c1 --parallel thread:32 --startres 30 --localrefine --iters=p3 --keep=0.95 --threads=32 --mask mask_tail3.hdf |
| Line 305: | Line 288: |
| e2spt_trajfromrefine.py --ali3dold spt_09/aliptcls3d_01.lst --ali3dnew spt_09/aliptcls3d_03.lst --maxshift 25 --nstd 3 --ali2d spt_09/aliptcls2d_03.lst --path spt_09 --nptcl 300 --nframe 8 --nbasis 1 }}} The program will compare the difference between the lst file from `--ali3dold` and `--ali3dnew` and build the eigen-trajectory from them. The 2D particle from `--ali2d` will define the reference frame of the movement. I.e. if you use aliptcls2d_01 instead of 03, you would see a rigid virus tail with a tilting membrane. You may need to filter the 3D volume stack to visualize the features properly. Simply run {{{ e2proc3d.py spt_05/threed_eig_00.hdf spt_05/threed_eig_00lp.hdf --process filter.lowpass.gauss:cutoff_freq=0.03 |
e2spt_trajfromrefine.py --ali3dold spt_04/aliptcls3d_01.lst --ali3dnew spt_04/aliptcls3d_04.lst --maxshift 25 --nstd 3 --ali2d spt_04/aliptcls2d_04.lst --path spt_04 --nptcl 300 --nframe 8 --nbasis 1 }}} The program will compare the difference between the lst file from `--ali3dold` and `--ali3dnew` and build the eigen-trajectory from them. The 2D particle from `--ali2d` will define the reference frame of the movement. I.e. if you use aliptcls2d_01 instead of 04, you would see a rigid virus tail with a tilting membrane. You may need to filter the 3D volume stack to visualize the features properly. Simply run {{{ e2proc3d.py spt_04/threed_eig_00.hdf spt_04/threed_eig_00lp.hdf --process filter.lowpass.gauss:cutoff_freq=0.03 |
| Line 323: | Line 306: |
| e2spt_mapptclstotomo.py --new --tomo tomograms/P22WT20210314A100__bin4.hdf --path spt_03 --iter 5 e2spt_mapptclstotomo.py --new --tomo tomograms/P22WT20210314A100__bin4.hdf --path spt_04 --iter 5 |
e2spt_mapptclstotomo.py --new --tomo tomograms/P22WT20210314A100__bin4.hdf --path spt_02 --iter 5 |
| Line 331: | Line 313: |
| It is also possible to map the full atom models instead of averaged structures back to the tomogram. A fancier version rendered with Unreal5 can be found [[https://drive.google.com/file/d/1xbpmEFKlWducYzGSdamtzKJ_CCdCcUAx/view|here]]. Tutorial for generating this maybe coming soon... |
|
| Line 333: | Line 317: |
| One limiting factor of the visualization of high resolution features in the tomograms is the subtle local misalignment. Since we corrected this during subtilt alignment, here we can apply those corrections back to the tomograms and improve the features inside. | One limiting factor of the visualization of high-resolution features in the tomograms is the subtle local misalignment. Since we corrected this during subtilt alignment, here we can apply those corrections back to the tomograms and improve the features inside. |
| Line 340: | Line 324: |
== Filament particle picking == Using the same dataset, we can also show some capabilities of filament refinement in the SPT workflow. From the tomograms, we can actually see two types of filament outside the cell, the bacterial flagella (blue) and pili (red). {{attachment:filament_in_tomo.png|filament in tomogram|width="300"}} Here we focus on the flagella. To pick the filament particles, from the '''e2tomo_eval.py''' window, select the target tomogram, then click '''Boxer''' while holding the '''Shift''' key. This will open a different particle picking interface for filament objects. Go up and down through the tomogram, and select curves by left clicking at the filament. The order of selection is generally irrelevant, and the program will still build the trace if you click the two ends of the filament first then modify the curvature by adding points at the center. To add a new curve (or select an existing different curve), hold '''Ctrl''' and click on the image. The program also allows selecting multiple types of filament by setting different '''ClassID'''. You can interpolate the curve with different spacing by clicking the '''Interpolate''' button, but it is only for visualization. After picking, close the small window. {{attachment:draw_curve.png|draw curve|width="500"}} == Filament particle extraction == To extract the filament particles, run {{{ e2spt_extract.py --boxsz_unbin=384 --newlabel=fiber0 --threads=8 --alltomograms --shrink=2.0 --tltkeep=1.0 --compressbits=8 --parallel=thread:4 --curves=0 --curves_overlap=.9 }}} Here instead of using `--label`, we use `--curve` to specify the type of filament particle we want to extract. The value of `--curve` is defined by '''ClassID''' in the curve drawing interface, and can be viewed in the tomogram evaluation window. `--curves_overlap` defines the overlap between neighboring boxes along a curve. i.e. `--curve_overlap=0` means the boxes do not overlap and 0.5 means each box overlaps with half of the next one. Here we use a large value to increase particle count since we do not have many filaments in the small dataset. Same as other particles, `--boxsz_unbin` defines the output box size. Since the curve drawing interface does not show boxes, it is a good idea to roughly measure the size of boxes using the regular manual boxer. The newly extracted particles will be labeled by `--newlabel`. From this dataset, I found 4 flagella filaments from 3 of the tomograms, and got 263 particles total. == Filament refinement == First, we make the initial model directly from the particles. {{{ e2spt_sgd_new.py sets/fiber0_bin2.lst --res 40 --curve }}} The `--curve` option will preserve the directional information from the curve and use that for the orientation assignment of particles. This should result in a reasonable initial model with some repeating features. {{attachment:filament_initial_model.png|filament initial model|width="600"}} The flagella have helical symmetry. Normally, we first refine the structure without symmetry, then estimate the symmetry from the subtomogram averaging results, and repeat the refinement with the correct symmetry. However, since we are limited by particle count, it is challenging to get anything with c1. So we simply take the konwn symmetry from existing single particle results from [[https://www.ebi.ac.uk/emdb/EMD-10362?tab=experiment|EMDB]]. Here we use `--sym h7:1:65.45:-1.15`, where `h7` making 7 symmetrical copies, `1` means no additional c symmetry, `65.45` is the helical turn in degrees, and `-1.15` is the rise in pixels. Here we add the minus sign before the rise value, since the convention in EMAN2 is different from that used in the EMDB entry. To confirm, download the EMDB map and apply the helical symmetry in EMAN2. To apply the symmetry, we still need to align the initial model to the symmetry axis. Since we use the information from the curve direction, the structure should already be quite close to the symmetry axis. To get to the correct axis, a simple way is to apply the symmetry first, then align the raw output to the symmetrical one. The process may need to be repeated a few times until it falls into the correct position. {{{ e2proc3d.py sptsgd_01/output_cls0.hdf sptsgd_01/output_cls0a.hdf --sym h7:1:65.45:-1.15 --process mask.soft:outer_radius=-40 e2proc3d.py sptsgd_01/output_cls0.hdf sptsgd_01/output_cls0b.hdf --align rotate_translate_3d_tree --alignref sptsgd_01/output_cls0a.hdf e2proc3d.py sptsgd_01/output_cls0b.hdf sptsgd_01/output_cls0a.hdf --sym h7:1:65.45:-1.15 --process mask.soft:outer_radius=-40 }}} Finally, we can do the refinement using the particles and the initial model. {{{ e2spt_refine_new.py --ptcls sets/fiber0_bin2.lst --ref sptsgd_01/output_cls0_xf_sym.hdf --sym h7:1:65.45:-1.15 --curve --parallel thread:32 --startres 30 --iters=p4,t2 --keep=0.95 --threads=32 --mask mask_cyl.hdf }}} Similar to the initial model, the `--curve` option will use the filament curve direction for orientation search. If the filament bends a lot, it is recommended to manually craft a shorter mask (using the '''mask.cylinder''' processor) to focus the alignment on a small segment. {{attachment:filament_refinement.png|filament refinement result|width="600"}} |
Advance tutorial of EMAN2 tomography workflow
This tutorial uses a CryoET dataset of P22 bacteriophage infecting cells, which is kindly provided by Jun Liu from Yale.
The dataset contains 4 tilt series and can be downloaded here.
This tutorial contains advanced topics of the SPT workflow, such as focused refinement and classification, symmetry breaking, and analysis of continuous motion. For new users, it is recommended to start with a simple tutorial.
Many of the functions here require an EMAN2 installation after 05/2023. A newer version is almost always better. A latest continuous build may work but compiling from source is preferred. For the installation guide, see here.
Create project folder
Make a new empty folder for the project and 'cd' into that folder.
Make sure all EMAN2 commands you run are executed from within this folder (not any subfolder).
Import tilt series
The files provided are already imported to EMAN2, so simply unzipping them in the project folder should work. For other CryoET projects, it is recommended to import the tilt series through e2projectmanager. Import tilt series will also convert them to hdf format, which greatly reduces storage (from 3.1GB to 700MB per tilt series) without obvious side effects.
After unzipping the files, you should have a folder called "tiltseries", with four hdf image stacks in it, inside the project folder. To view the tilt series, run e2display.py, locate the file in the browser, and click Show2D.
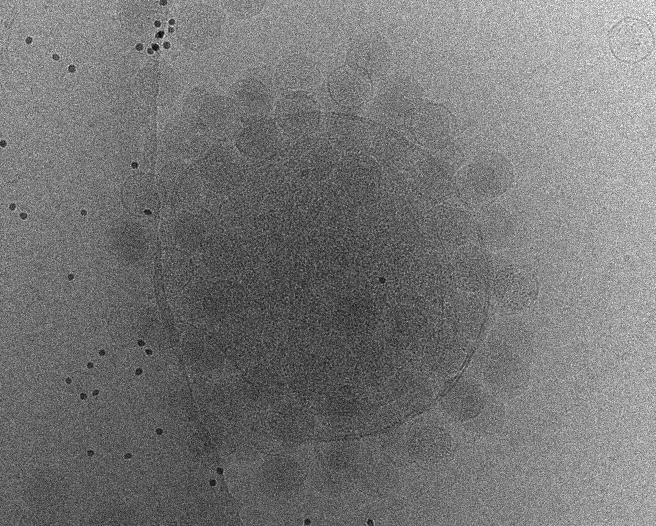
Initial tomogram reconstruction
Start from one good tilt series to play with the parameters for tilt series alignment and tomogram reconstruction. You often need to run this multiple times with different options to get a satisfying reconstruction. After having a good set of options, we will run the program with the same parameters for all tilt series later.
e2tomogram.py tiltseries/P22WT20210314A100.hdf --zeroid=-1 --tltstep=3.0 --npk=20 --tltkeep=0.9 --outsize=1k --niter=2,1 --bytile --pkkeep=0.9 --compressbits=8 --clipz=320 --bxsz=32 --filterres=60.0 --rmbeadthr=-1.0 --threads=12 --patchtrack=2 --autoclipxy
- Always check --autoclipxy for non-square micrographs. The program will decide how to clip based on the content of the tomogram.
- Patchtracking (--patchtrack=2) now works almost as well as landmark tracking (controlled by --niter=X) when the fiducials are not present. It should not stop you from getting to ~3Å with enough particles and good samples, even with --niter=0. Having both normally does not hurt.
- Tomogram thickness needs to be decided visually. The default often works, but it is good to increase the number if there are still features at the top and bottom of the tomogram.
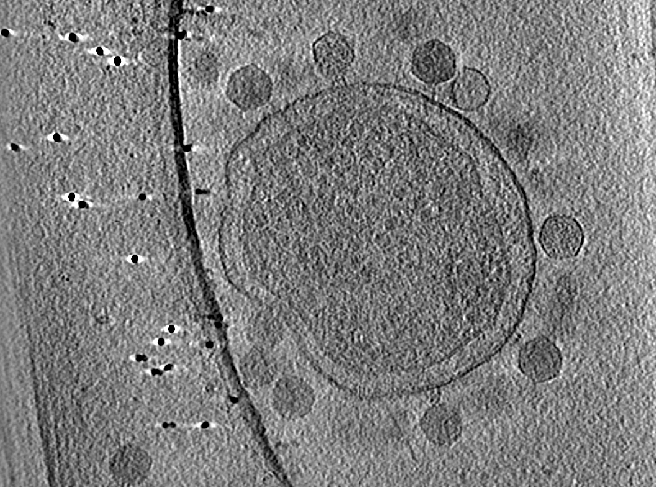
Handedness check by CTF
Before reconstructing all tilt series, we need to make sure the handedness of the tomogram is correct.
e2spt_tomoctf.py tiltseries/P22WT20210314A100.hdf --dfrange=2.0,7.0,0.02 --psrange=10,15,5 --tilesize=400 --voltage=300 --cs=2.7 --nref=15 --stepx=20 --stepy=40 --checkhand --threads=1 --writetmp
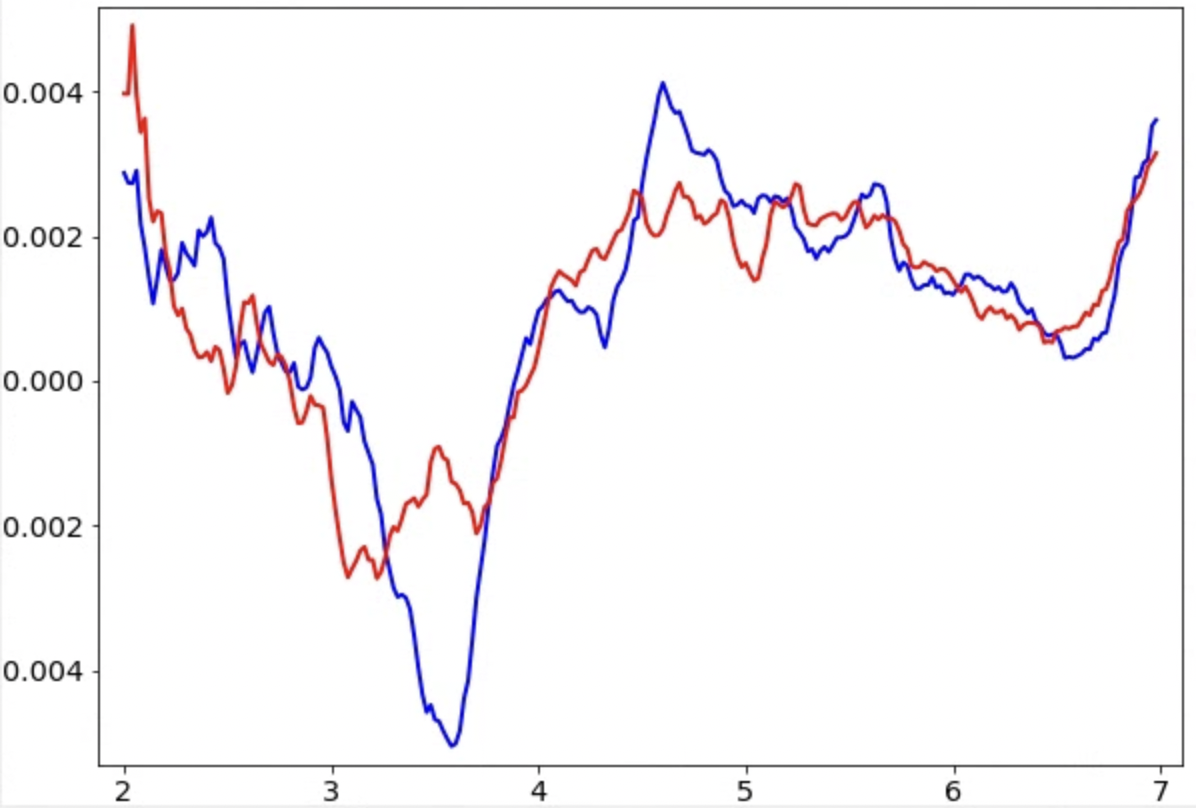
The program will print out the recommended tilt axis angle from the command line, and we will reconstruct all tomograms using the new tilt axis angle (--tltax option in e2tomogram). The result relies on the fitting of CTF rings in the micrographs, and can be unreliable if there is not enough signal. Only believe the result when >80% of the tilt angles have the same handedness. It is better to also check the temporary files, which are written in tomoctf_xx if --writetmp is specified. At high tilt, i.e. tilt ID close to 0 or the 35, the curve of one of the handedness (blue) should show a much deeper dip than the other one (red). If it is not obvious, something in the earlier steps might be wrong and the images might not have enough signal for CTF estimation. Also, if the dip is too close to the left or right end of the plot, consider adjusting the range of defocus search using --dfrange.
Note this only accounts for the geometry of the tilt series, but it can still produce the wrong handedness if your individual micrographs are flipped. This can sometimes be the case with some data collection software. Even in those cases, you should still use the handedness recommended by the program (or go back and flip the raw micrographs), which will produce a more accurate defocus estimation.
All tomogram reconstruction
Reconstruct all tilt series using the same parameters and the tilt axis estimated by the handedness check.
e2tomogram.py --alltiltseries --zeroid=-1 --tltstep=3.0 --npk=20 --tltax=-4.2 --tltkeep=0.9 --outsize=1k --niter=2,1 --bytile --notmp --pkkeep=0.9 --compressbits=8 --clipz=320 --bxsz=32 --filterres=60.0 --moretile --rmbeadthr=-1.0 --threads=12 --patchtrack=2 --autoclipxy
CTF estimation
Now we estimate the defocus for all tilt series. Note here we only estimate the CTF, not correct for it. The correction will happen later when extracting particles. To get to high resolution, it is often safer to use a higher than the default --tilesize.
e2spt_tomoctf.py --alltiltseries --dfrange=1.5,7.0,0.02 --psrange=10,15,5 --tilesize=400 --voltage=300 --cs=2.7 --nref=15 --stepx=20 --stepy=40 --threads=4
Evaluate tomograms
One convenient way to go through many tomograms in a dataset is the tomogram evaluation GUI. To use it, launch from the project manager or run
e2tomo_eval.py
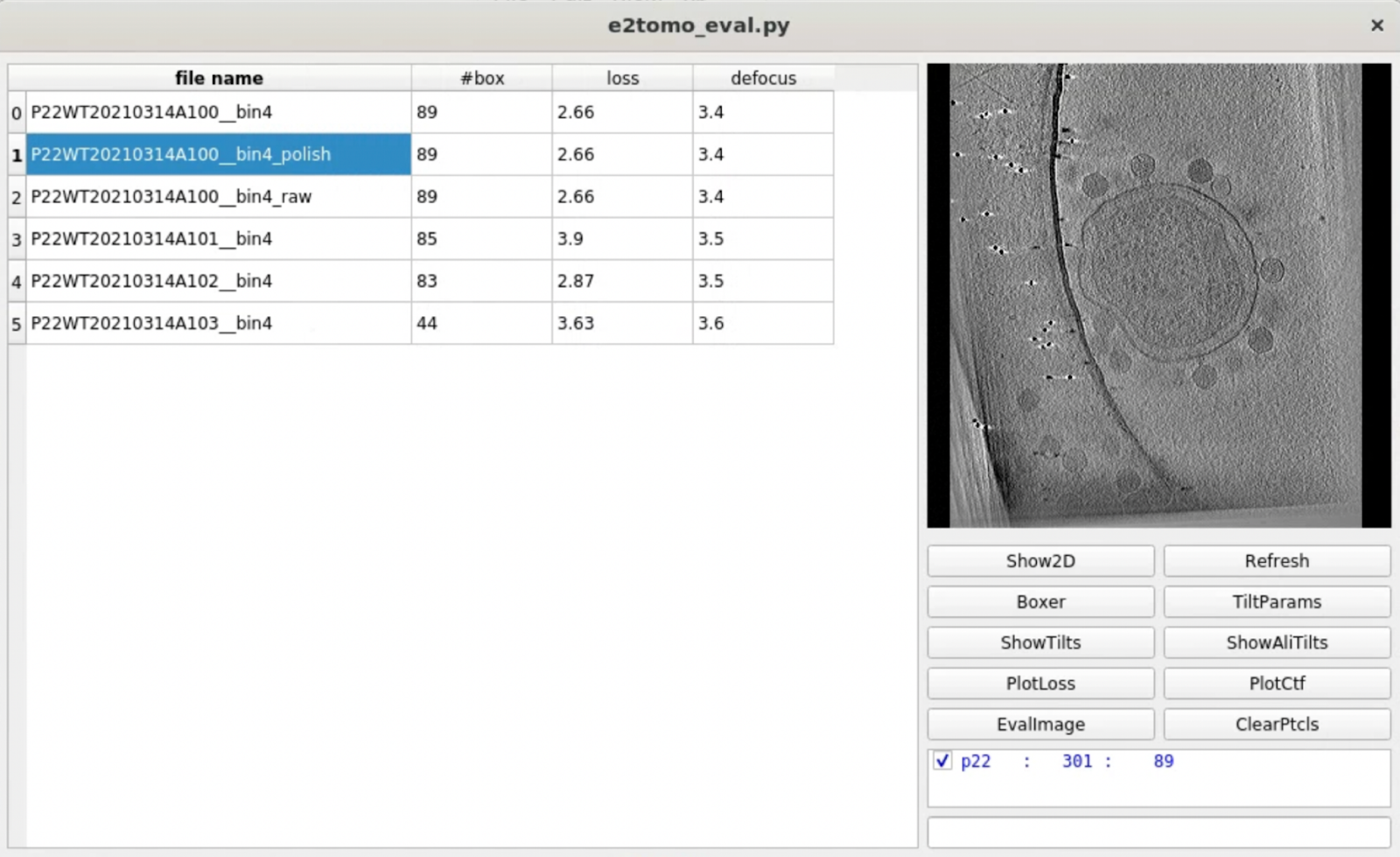
You can visualize tomograms, raw and aligned tilt series, as well as related alignment parameters from the GUI. Manual particle boxing can also be launched from the tool.
Convolutional network based particle picking
In this tutorial, we select particles using the automatic particle picker. This program is deep neural network based, and requires GPU and CUDA installed. Without CUDA, it is also possible to manually pick a few particles, generate initial model, then use that model for template matching. It should work almost as well since the virus is an easy target. Here we run
e2spt_boxer_convnet.py --label=p22
Since the particles are quite large, we change the reference box size to 192 using the ChangeBx button first. Start from 5 positive and 5 negative references, train and apply the network, then refine the network by adding more false positives/negatives from multiple tomograms to the training set. Apply the trained network to all tomograms then close the window. Here we get ~300 particles from 4 tomograms.
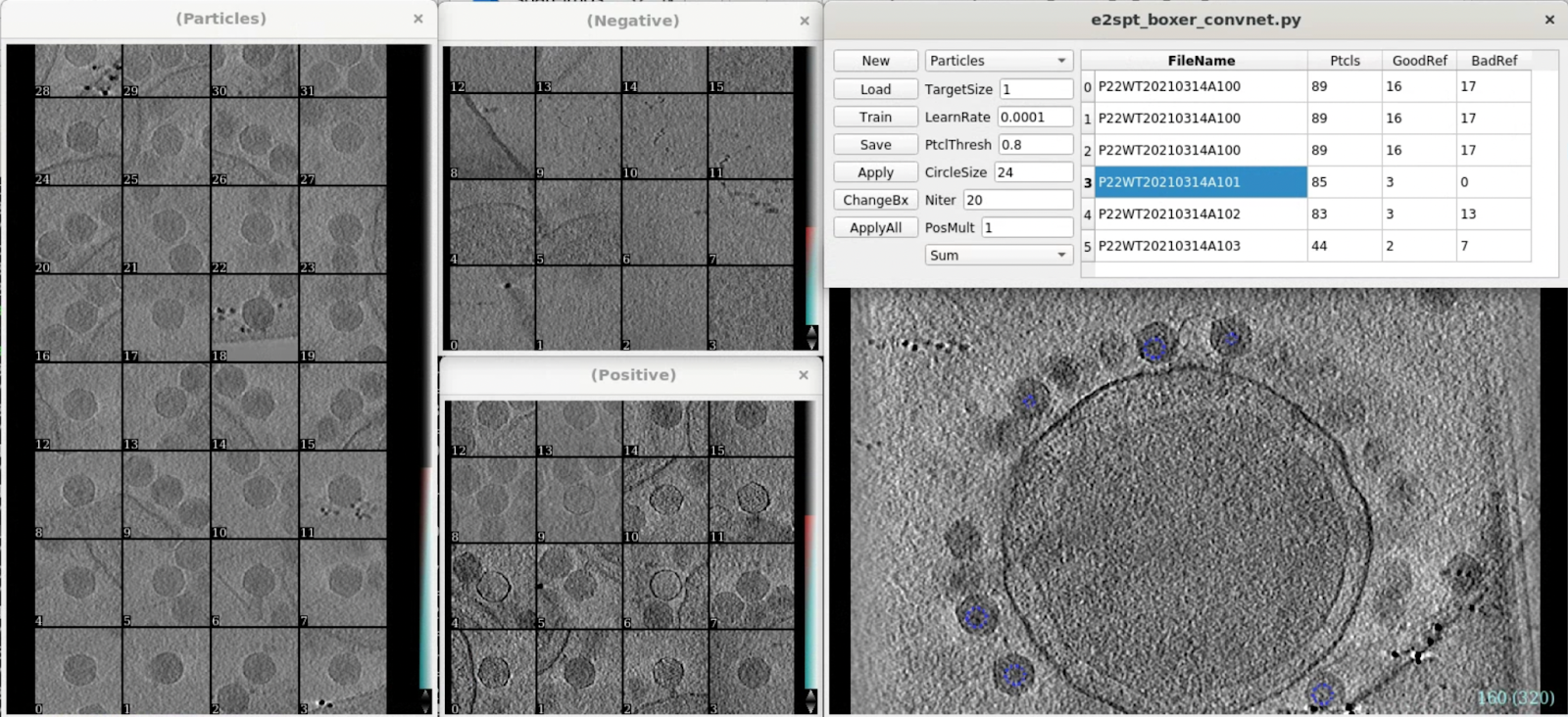
After auto-boxing, we can take a look at the particles using e2spt_boxer.py to make sure they are correct, and also decide the correct box size.
Particle extraction
After selecting all particles, we extract particles of the P22 capsids. For speed and memory consumption, we use a box size of 640 and shrink the particles by 4 using --shrink. After extraction, run e2spt_buildstack.py to generate a virtual stack of all particles of the same time from different tomograms.
e2spt_extract.py --boxsz_unbin=640 --label=p22 --threads=8 --alltomograms --shrink=4.0 --tltkeep=1.0 --compressbits=8 --parallel thread:4 e2spt_buildsets.py --allparticles
Refinement workflow
Initial model generation
We start by generating the initial model directly from the particles. Since the icosahedral capsid does not have many low-resolution features, here we set --sym=icos and --res=20 to target a higher resolution.
e2spt_sgd_new.py sets/p22_bin4.lst --res=20.0 --niter=100 --shrink=1 --parallel=thread:17 --ncls=1 --batch=12 --learnrate=0.2 --sym=icos
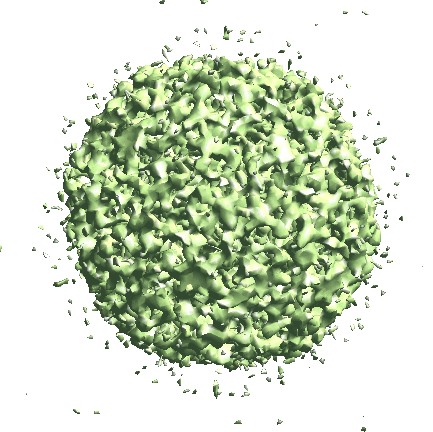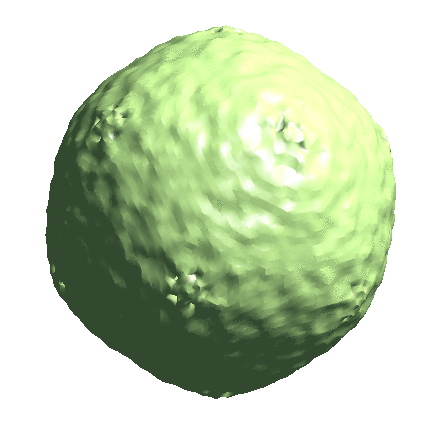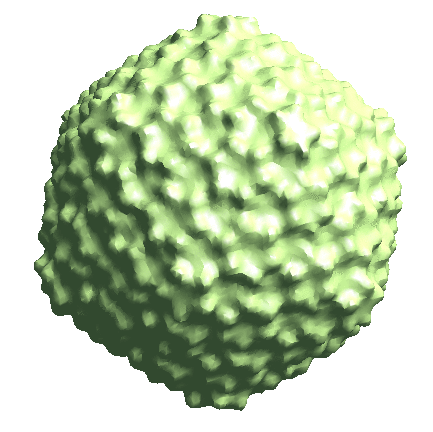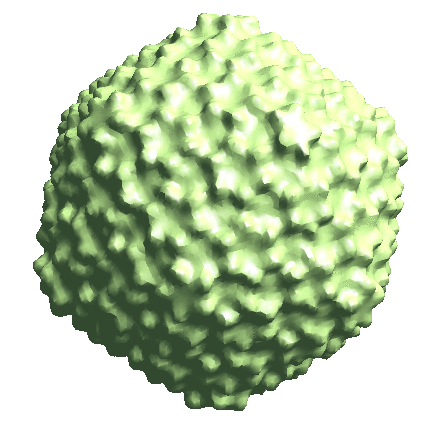
Depending on the quality of particles, there can be some randomness in the initial model generation process, and the command can sometimes converge to a featureless sphere. A safer option is to set --ncls=3, so the program will generate 3 initial models at the same time, but this will also be slower.
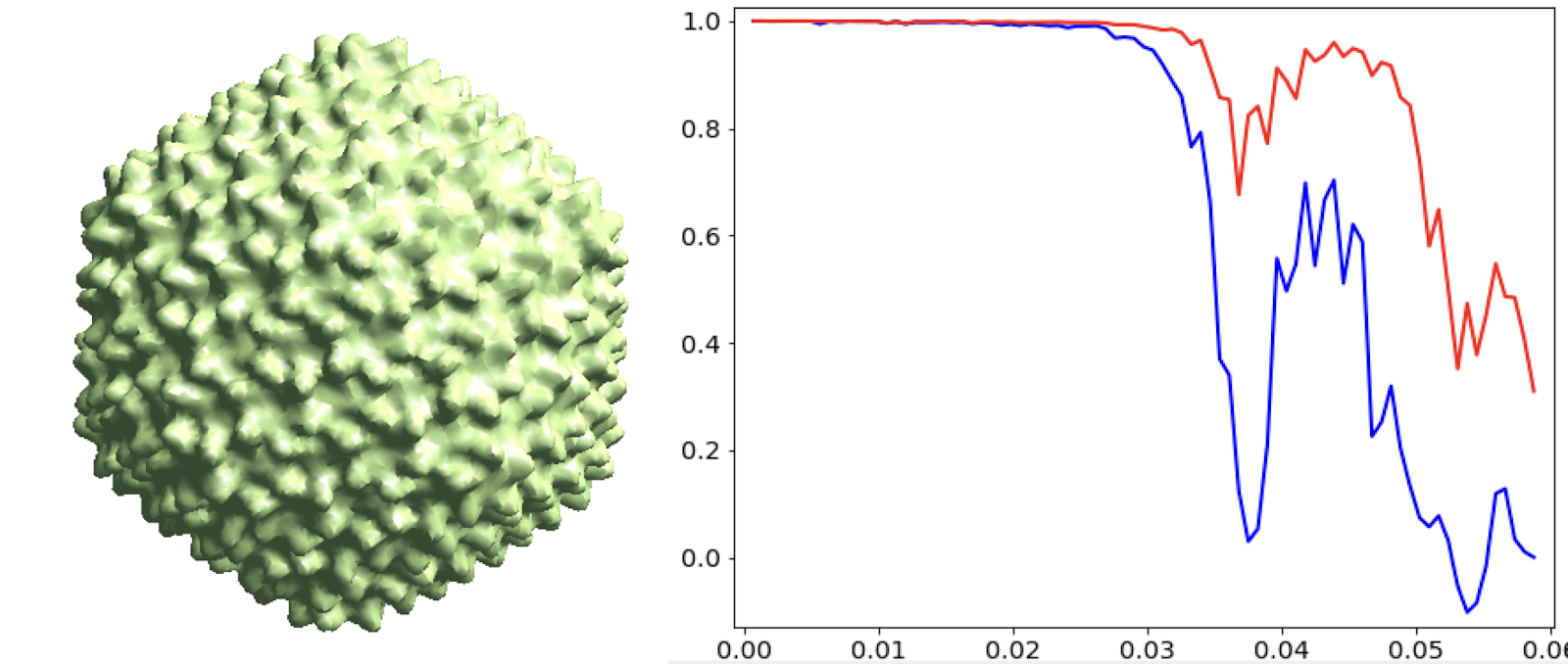
Icosahedral capsid refinement
Start from the symmetrical refinement of the capsid. Here we use the "new" refinement routine in e2spt_refine_new.py that integrates the subtomogram (p iterations) and subtilt refinement (t and r iterations). The refinement should reach Nyquist of the particles (17Å) easily. One thing to note here is the FSC curve should improve after the subtilt refinement (red vs blue curve below). If it does not, it means either the data does not have enough signal per particle per tilt for the alignment, which may greatly limit the resolution achieved, or the tilt series alignment is so good that there isn’t much to improve, which does not really happen often.
e2spt_refine_new.py --ptcls=sets/p22_bin4.lst --ref=sptsgd_00/output_cls0.hdf --startres=30.0 --goldstandard --sym=icos --iters=p3,t2,p,t,r,d --keep=0.95 --parallel=thread:32 --threads=32
Finding tail using symmetry breaking
After the icosahedral capsid refinement, we have two main goals here. One is to push the symmetrical capsid structure toward a higher resolution, and the other is to find the tail of the virus and solve its structure. However, the two goals are somewhat intertwined, and we will need to get back and forth a bit to solve both. Here we first look for the tail of the phage with symmetry breaking. Start by running e2spt_refinemulti_new.py targeting a low resolution (--maxres=80).
e2spt_refinemulti_new.py --nref 1 --ptcls spt_00/aliptcls3d_06.lst --niter 5 --maxres 80 --breaksym icos --loadali3d --parallel thread:32 --sym c5 --skipali --loadali2d spt_00/aliptcls2d_08.lst
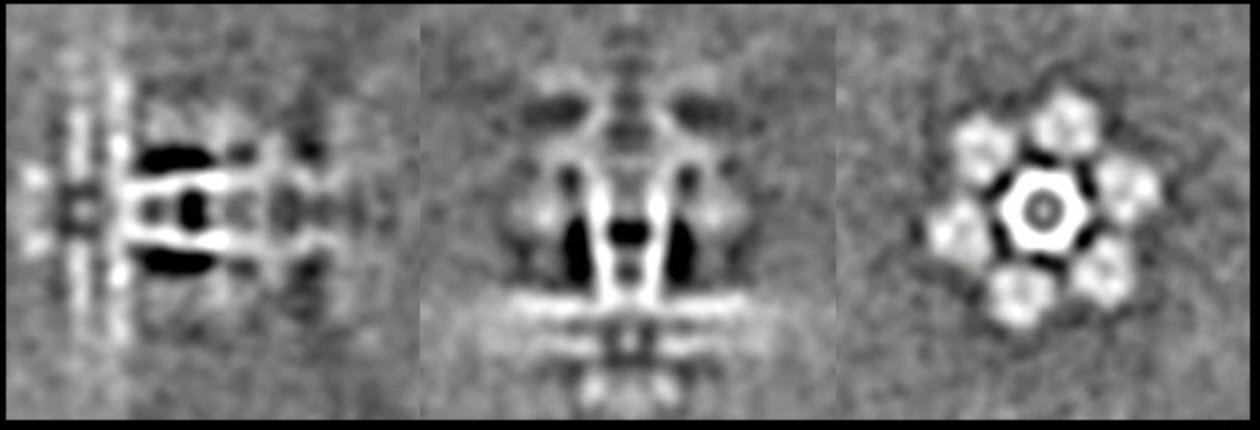
Here we load both the 3D subtomogram orientation assignment and subtilt alignment from the previous refinement folder. Since we break the icosahedral symmetry and re-apply the c5 symmetry, the densities of the tail should show up the penton at either the top or the bottom of the capsid after a few iterations. One easy way to visualize this is to look at the central slice views. In the e2display browser, select the map and click RngXYZ. Here we set Sum layers to 10 to look at a 20-pixel thick slice.
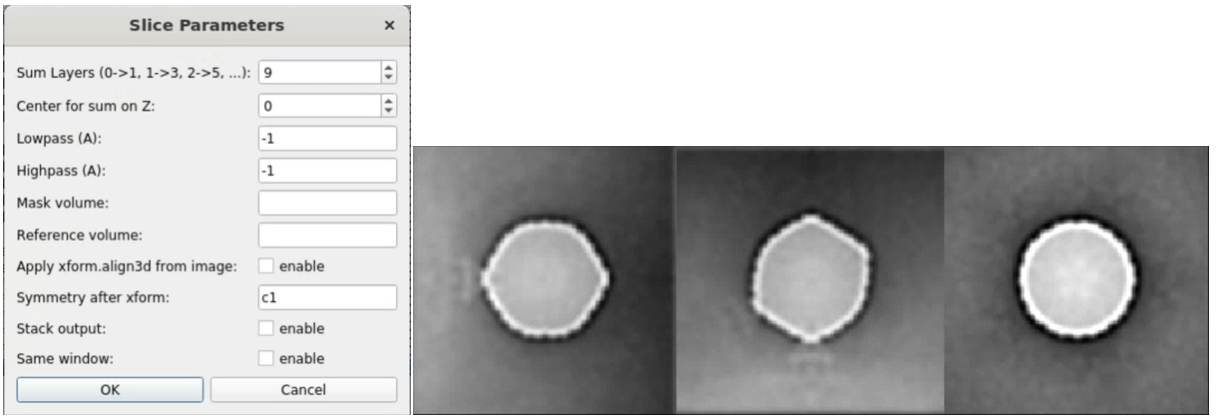
Now we have an initial capsid structure with a tail, we can do another run of the symmetry breaking using this as the reference, and target a higher resolution. To ensure convergence, we would like to craft a mask that covers only the location of the tail we have in the reference. It is more convenient to make the mask in Filtertool, which can be launched by selecting the reference map (sptcls_00/threed_05_00.hdf), and clicking Filtertool in the e2display browser. If you have a slow machine and the program crashes often, hold Shift and click the button to enter safe mode. In the Fitertool, first add a soft mask using mask.soft, and change outer_radius and dz so it covers only the density of the tail. Afterward, add a math.linear processor before the soft mask, which will set the map to all ones, so the output would be a spherical mask with a soft edge instead of masked-out density. Save the mask using File->Save, then rename it to mask_tail0.hdf. Finally, we add the mask of the capsid to the mask of the tail to make sure the alignment does not drift away from the initial capsid refinement.
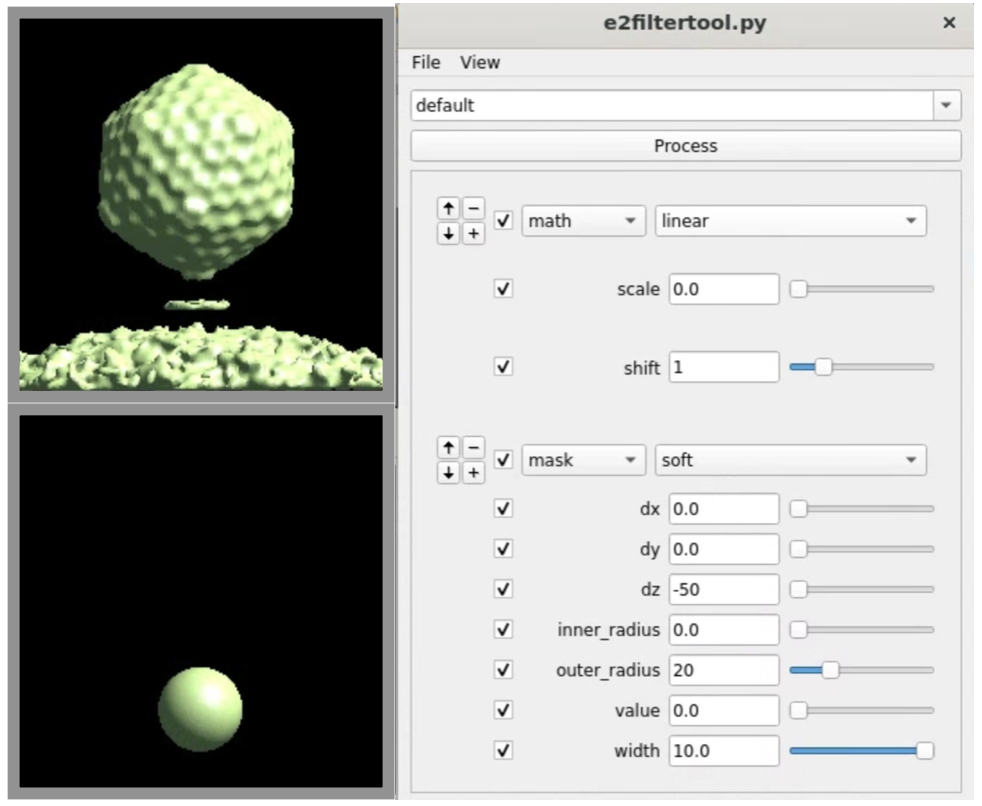
e2proc3d.py mask_tail0.hdf mask_tail1.hdf --addfile spt_00/mask.hdf --process threshold.clampminmax:maxval=1
Here we also add a threshold.clampminmax processor to make sure there is no value above 1 in the mask.

With the mask ready, we can now start the second symmetry breaking run. The program will be slower since we took out the --skipali option, and will do local orientation refinement in addition to symmetry breaking.
e2spt_refinemulti_new.py sptcls_00/threed_05_00.hdf --ptcls spt_00/aliptcls3d_06.lst --niter 10 --maxres 30 --breaksym icos --loadali3d --parallel thread:32 --minres 150 --sym c5 --maskali mask_tail1.hdf --loadali2d spt_00/aliptcls2d_08.lst

Since we are applying the c5 symmetry here, the tail will not be fully resolved, but the location of the tail should be mostly correct, which is indicated by the solid density in the structure after the refinement.
Now that we found the tail of each capsid, we can re-extract the unbinned particles of the tail base on the refinement results. In particle extraction, ignore the tomogram options, and only input the aliptcles3d_xx.lst through the --jsonali option. The program will find the particles given the refinement results. The program also allows extracting particles with an offset from the previous refinement, so here we can specify --postxf=icos,0,0,-50, where (0,0,-50) is the coordinates of the tail from the averaged structure. At this point, we will only extract the particles and build sets (this will give us sets/tail.lst), but do not refine the tail structure yet.
e2spt_extract.py --boxsz_unbin=208 --newlabel=tail --threads=8 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=sptcls_01/aliptcls3d_10_00.lst --compressbits=8 --postxf c1,0,0,-50 --mindist 100 --parallel thread:4 e2spt_buildsets.py --allparticles --label tail
Penton refinement
Now we get back to the icosahedral capsids and extract particles at each penton of the capsid to refine them locally. The particle extraction program can generate multiple new particles from each original one with a given symmetry. Here we want to get all 12 pentons for each capsid, so we specify --postxf=icos,0,0,35, where (0,0,35) is the coordinates of one penton from the capsid refinement. Make sure to also set a number for --mindist, which defines the minimum distance between the new particles. Here the program will initially find 60 new penton particles for each capsid particle based on the icosahedral symmetry, then reduce the number to 12 since every 5 particles would have the same coordinates.
e2spt_extract.py --boxsz_unbin=192 --newlabel=pent --threads=8 --maxtilt=100 --padtwod=2.0 --shrink=1 --tltkeep=1.0 --jsonali=spt_00/aliptcls3d_03.lst --compressbits=8 --postxf icos,0,0,35 --mindist 100 --parallel thread:4 e2spt_buildsets.py --allparticles --label pent
After extraction, instead of starting a refinement directly, we first gather the metadata of the newly extracted particles using the e2spt_gathermeta.py program. The program will collect the information from particle headers so the new particles inherit the orientation and even/odd split of the previous refinement. With the --ali2d input, it also interpolates the subtilt movement of each new 2D particle from the movement of particles in the same tilt from the previous alignment.
Also note that out of the 12 pentons in each capsid, the penton with the tail would have a different structure than the others. Out of an abundance of caution, we do not use all pentons we just extracted, but remove the ones that are close to the tails we found earlier. This is done by providing the tail particles through the --exclude option, which is why we need to find the tails before this step. In practice, since it is only 1/12 of the entire population, the pentons with tails will likely be excluded or down-weighted in the iterative refinement, and won't actually cause any difference. But it is still a safer practice to avoid potential issues down the way.
e2spt_gathermeta.py --ptcls sets/pent.lst --ali2d spt_00/aliptcls2d_05.lst --exclude sets/tail.lst
Now run the refinement in the same folder (spt_01) continuing from the metadata we just gathered. Here --continuefrom=0.5 will do reconstruction of iteration 1, followed by alignment of iteration 2. Since we keep the same even/odd split from the capsid refinement and particles from the two subsets still have not seen each other, we can omit the --goldstandard option and simply continue the refinement.
e2spt_refine_new.py --path spt_01 --continuefrom 0.5 --sym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --iters=p2,t2,p,t,r,d --keep=0.95 --threads=32
This should get better than 7Å at the end. At this point, the SSEs from the model (PDB: 5UU5) can fit in nicely.
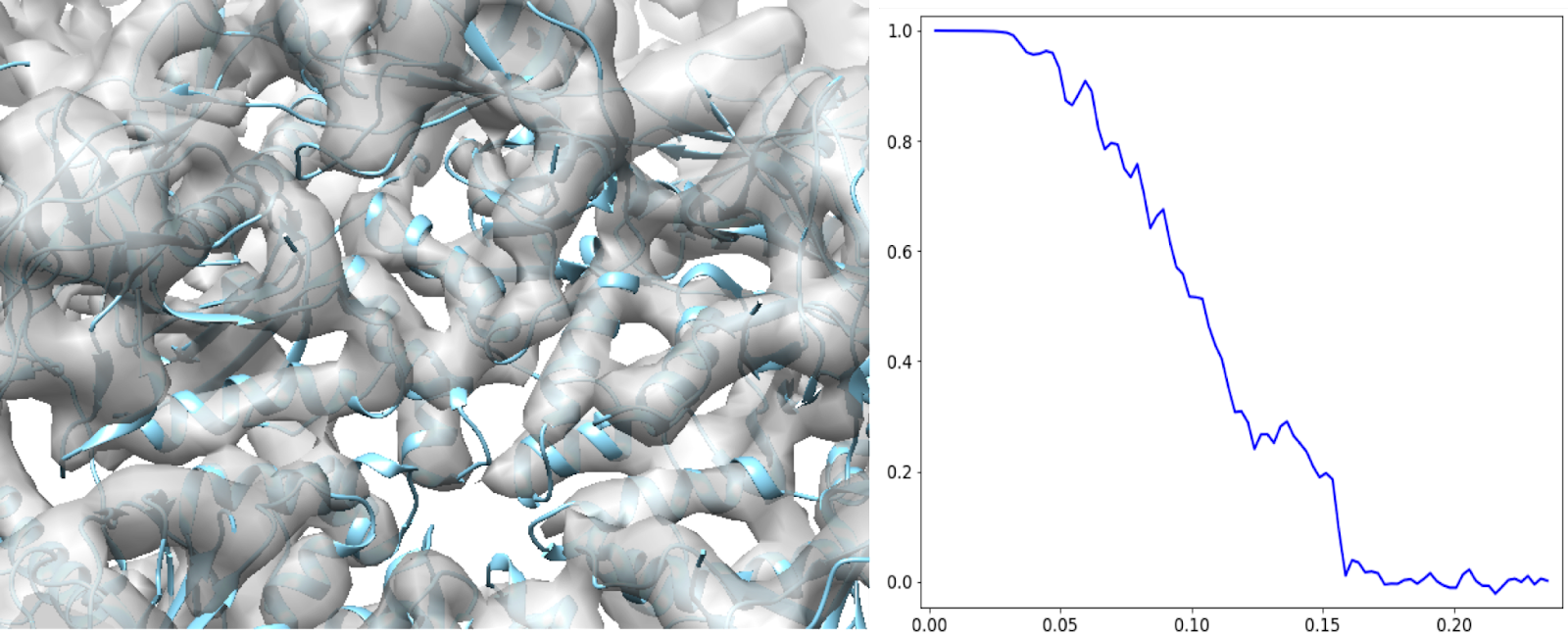
Evaluating subtilt refinement
After the refinement (or at least one t iteration) is finished, we can run the following command to evaluate the results of a subtilt refinement.
e2spt_evalsubtlt.py --path spt_01
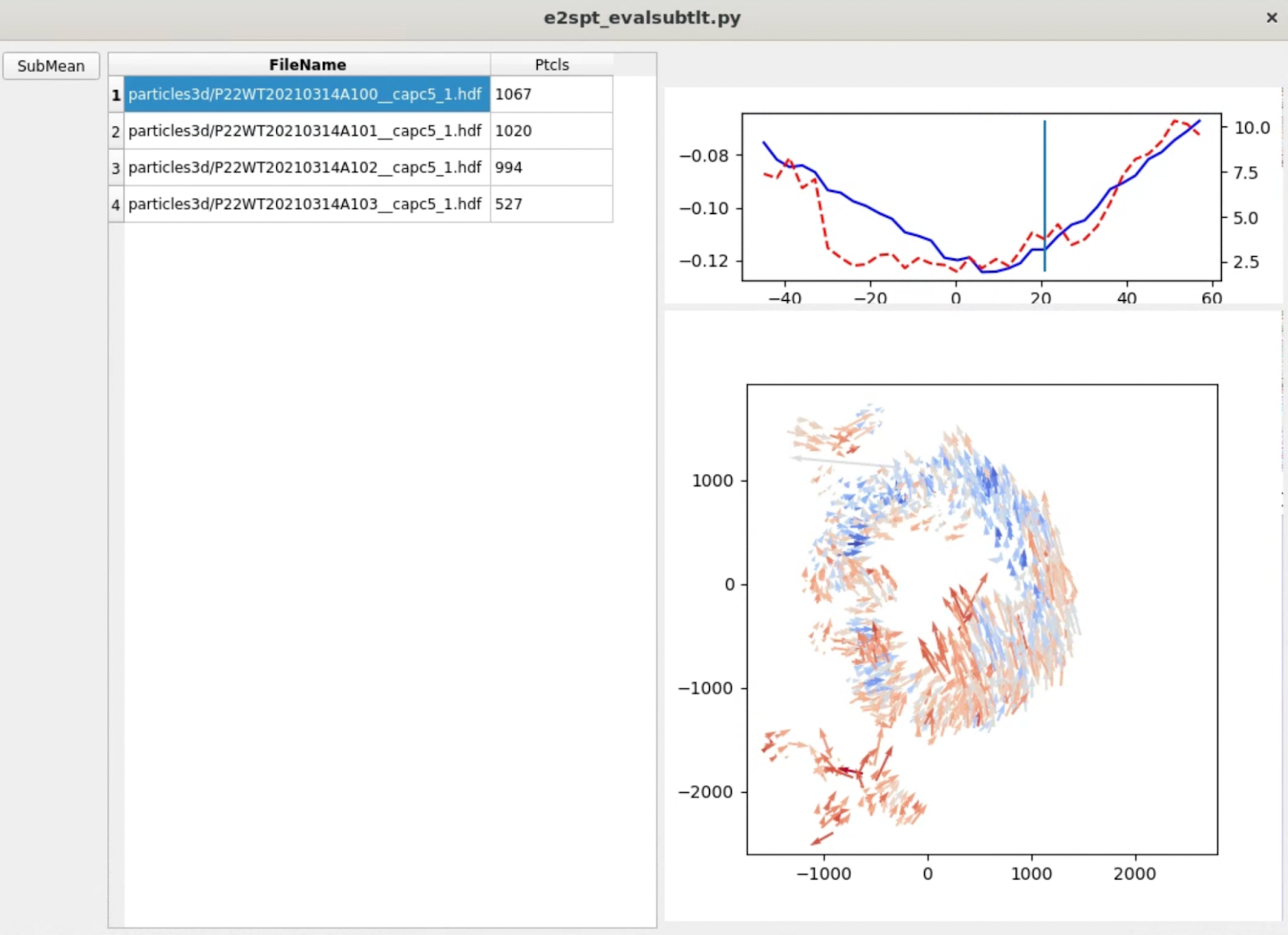
Typically, you should see the amount of movement in each tilt (red dashed line) roughly correlated with the particle score (blue line), both of which increase at higher tilt. In each tilt, the vectors should form a somewhat locally correlated flow field. Note the length of arrows is exaggerated, since they normally do not move at a length that’s visible at the scale of the entire tomogram.
Tail refinement
Now back to the tail refinement. Since we now have the high resolution of the capsid pentons, we will use the subtilt alignment information from there to guide the refinement of the tail. But first, we still need to break the c5 symmetry we imposed previously and re-apply the c6 symmetry, to get an initial structure of the tail.
e2spt_refinemulti_new.py --nref 1 --ptcls sets/tail.lst --niter 10 --maxres 30 --breaksym c5 --loadali3d --parallel thread:32 --minres 200 --sym c6 --skipali

Before refining the tail, we would like to make a customized mask for it, instead of using the auto masking routine in EMAN2. The goal here is to remove the impact of the capsid density which has c5 symmetry to prevent it from impacting the alignment. Again, we make this using the Filtertool. Here we also start from a mask.soft processor to get to rough location of the tail, then add a mask.auto3d.thresh processor after it to generate the mask. This processor selects all voxels at threshold2 that are connected to the voxels above threshold1, and adds extra padding afterward. We rename this mask mask_tail1.hdf.
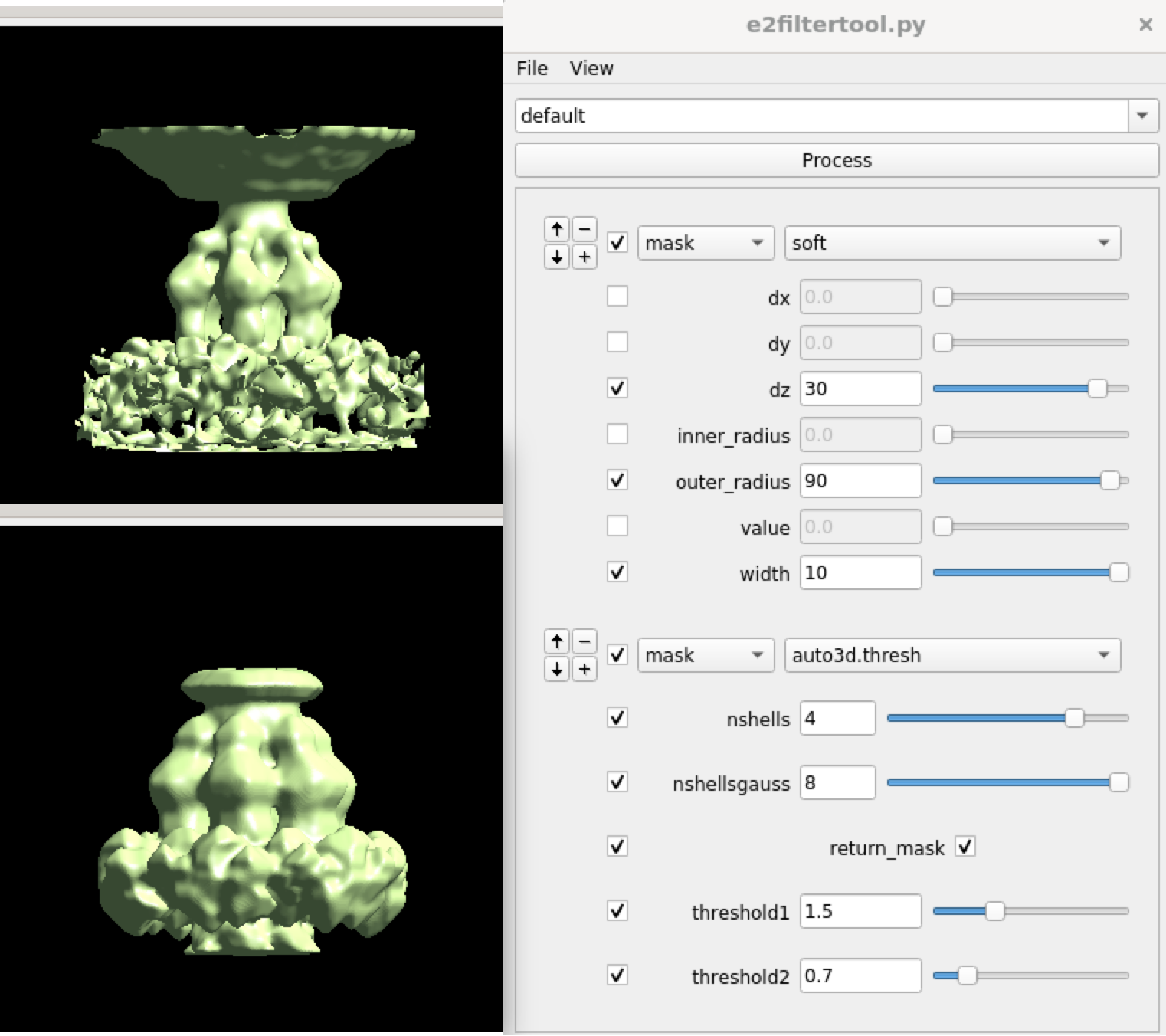
After this, we can start the refinement. First, interpolate the subtilt movement of the capsid and apply it to the nearby tail particles. Then locally refine the structure of the tail. Here we did symmetry breaking at the e2spt_refinemulti_new.py as well as e2spt_refine_new.py. It is actually okay to skip the symmetry breaking in the second refinement, but it does not hurt to do an extra round just to be safe.
e2spt_gathermeta.py --ptcls sptcls_02/aliptcls3d_10_00.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d
This creates spt_02 and compiles metadata inside. Then,
e2spt_refine_new.py --path spt_02 --continuefrom 0.5 --sym c6 --breaksym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --iters=p3,t2,p,t --keep=0.95 --threads=32 --mask mask_tail2.hdf

Further classification of the tail
The single model refinement will get to ~15Å resolution. One limitation of the refinement is the heterogeneity at the lower part of the tail where it interacts with the membrane. This is likely because only some of the particles are anchored on the membrane, whereas the others are floating in the solution. To sort them out, do some more classification. Similarly, we create a soft mask that covers the lower part of the tail, and call it mask_tail3.hdf
e2spt_refinemulti_new.py --nref 2 --ptcls spt_02/aliptcls3d_07.lst --niter 10 --maxres 25 --loadali3d --parallel thread:32 --minres 200 --sym c6 --maskali mask_tail3.hdf --loadali2d spt_02/aliptcls2d_08.lst --skipali
This will yield two classes, one with tails floating, and the other with tails sitting on the membrane.
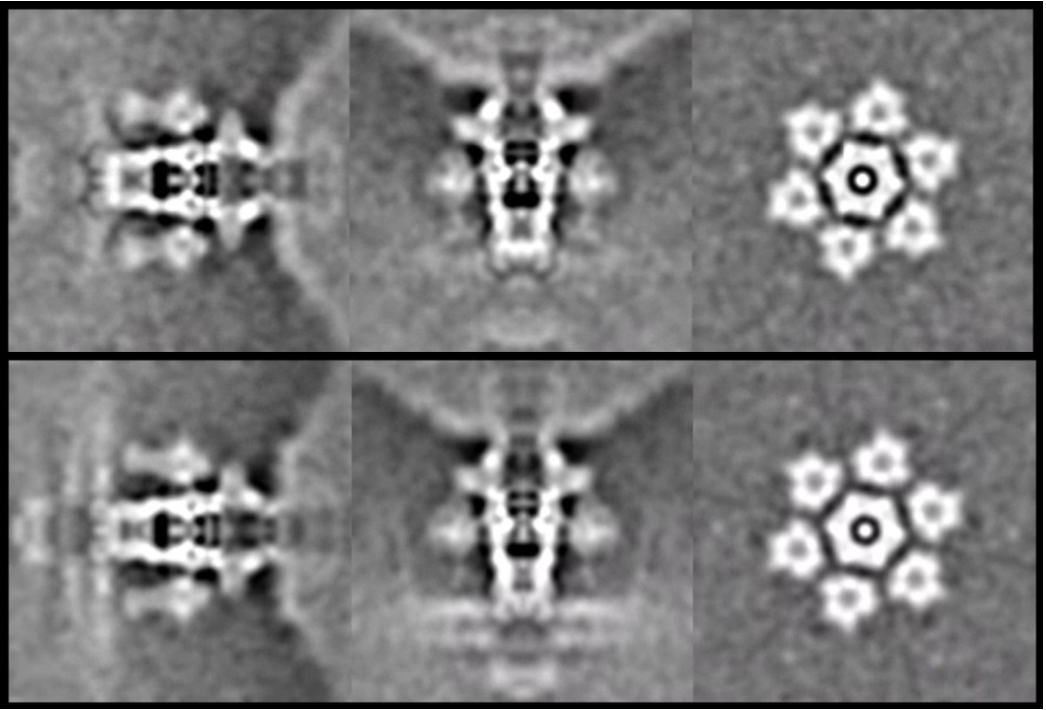
It is also possible to further refine the structure of individual classes. This will give us better features at the membrane channel, but the resolution will be limited by particle count.
e2spt_gathermeta.py --ptcls sptcls_03/aliptcls3d_10_01.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d e2spt_refine_new.py --path spt_03 --continuefrom 0.5 --sym c6 --breaksym c5 --parallel thread:32 --startres 20 --tophat localwiener --localrefine --iters=p2,t2,p,t --keep=0.95 --threads=32 --mask mask_tail2.hdf
Continuous movement
There are probably multiple modes of continuous movement within the system, but most of them are tricky to deal with since we are very low on particle count at this moment. Here we focus on a relatively simple example, the overall tilting of the virus tail with respect to the membranes.
First, expand the symmetry of the particles, and make 6 copies of each tail particle at the symmetrical orientations. e2proclst.py only expands the symmetry in place so we need to make a copy of the file first.
e2proclst spt_03/aliptcls3d_05.lst --create spt_03/aliptcls3d_05_c6.lst e2proclst.py spt_03/aliptcls3d_05_c6.lst --sym c6
Then do a few rounds of refinement focusing on the membrane and density below.
e2spt_gathermeta.py --ptcls spt_03/aliptcls3d_03_c6.lst --ali2d spt_01/aliptcls2d_05.lst --ali3d e2spt_refine_new.py --path spt_04 --continuefrom 0.5 --sym c1 --parallel thread:32 --startres 30 --localrefine --iters=p3 --keep=0.95 --threads=32 --mask mask_tail3.hdf
To make sure the focused refinement works properly, take a look at the unmasked results in spt_xx/threed_raw_even/odd.hdf. Here we should see solid density of the membrane and channel, with vague densities of the virus tail above.
Finally, we can compile the trajectory of the continuous movement by comparing the difference of angle assignment between the two refinements.
e2spt_trajfromrefine.py --ali3dold spt_04/aliptcls3d_01.lst --ali3dnew spt_04/aliptcls3d_04.lst --maxshift 25 --nstd 3 --ali2d spt_04/aliptcls2d_04.lst --path spt_04 --nptcl 300 --nframe 8 --nbasis 1
The program will compare the difference between the lst file from --ali3dold and --ali3dnew and build the eigen-trajectory from them. The 2D particle from --ali2d will define the reference frame of the movement. I.e. if you use aliptcls2d_01 instead of 04, you would see a rigid virus tail with a tilting membrane. You may need to filter the 3D volume stack to visualize the features properly. Simply run
e2proc3d.py spt_04/threed_eig_00.hdf spt_04/threed_eig_00lp.hdf --process filter.lowpass.gauss:cutoff_freq=0.03
Then display the movie by clicking the Show All 3D button.
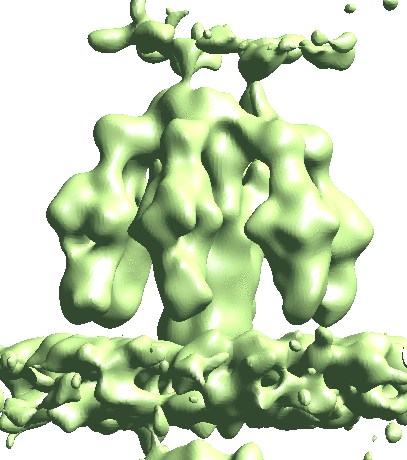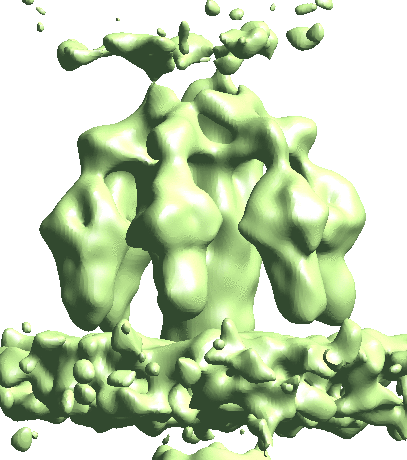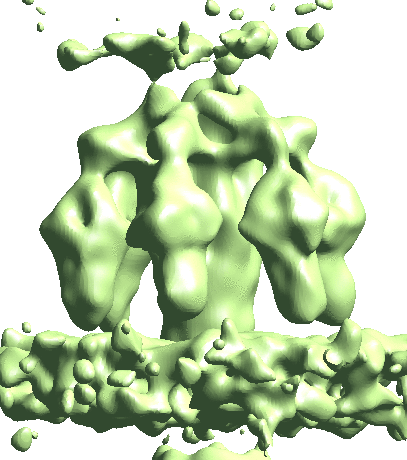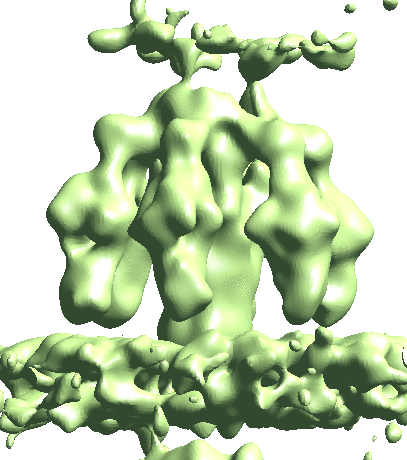
Map particles to tomograms
After refinement, we can map particles back to the original tomograms to visualize their organization inside the tomogram. We can also do this for multiple refinements and visualize them in the same tomogram.
e2spt_mapptclstotomo.py --new --tomo tomograms/P22WT20210314A100__bin4.hdf --path spt_00 --iter 3 e2spt_mapptclstotomo.py --new --tomo tomograms/P22WT20210314A100__bin4.hdf --path spt_02 --iter 5
Then open the raw tomogram and the mapped back particles (spt_00/ptcls_in_tomo_P22WT20210314A100_03.hdf etc.) in Chimera.

It is also possible to map the full atom models instead of averaged structures back to the tomogram. A fancier version rendered with Unreal5 can be found here. Tutorial for generating this maybe coming soon...
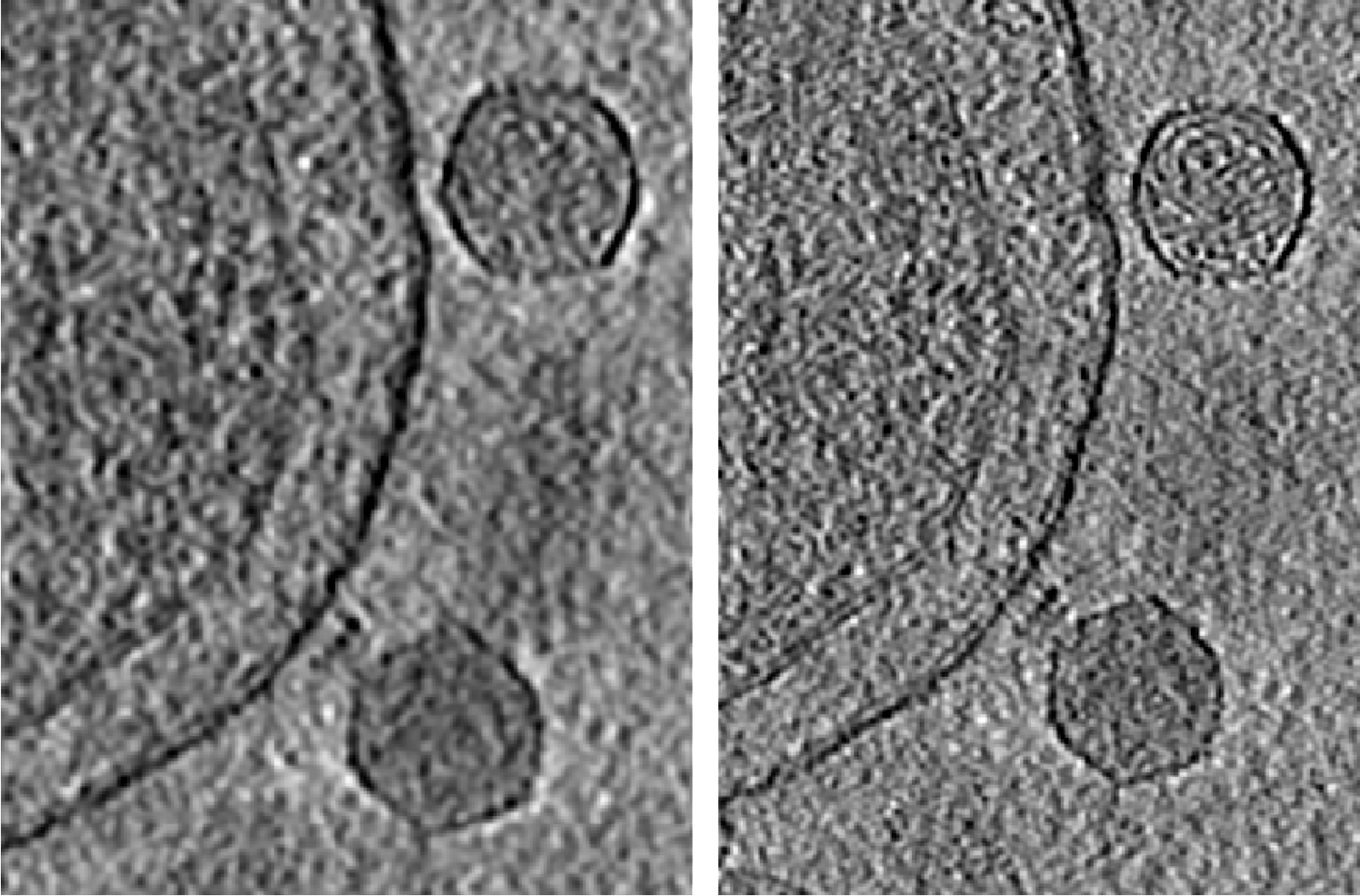
Polish tomogram using subtilt refinement
One limiting factor of the visualization of high-resolution features in the tomograms is the subtle local misalignment. Since we corrected this during subtilt alignment, here we can apply those corrections back to the tomograms and improve the features inside.
e2spt_polishtomo.py --path spt_00 --fname tomograms/P22WT20210314A100__bin4.hdf --res 40
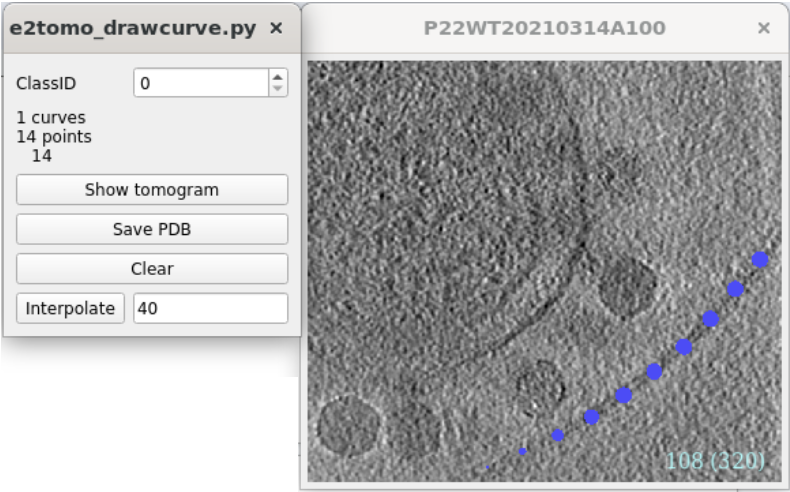
Filament particle picking
Using the same dataset, we can also show some capabilities of filament refinement in the SPT workflow. From the tomograms, we can actually see two types of filament outside the cell, the bacterial flagella (blue) and pili (red).

Here we focus on the flagella. To pick the filament particles, from the e2tomo_eval.py window, select the target tomogram, then click Boxer while holding the Shift key. This will open a different particle picking interface for filament objects. Go up and down through the tomogram, and select curves by left clicking at the filament. The order of selection is generally irrelevant, and the program will still build the trace if you click the two ends of the filament first then modify the curvature by adding points at the center. To add a new curve (or select an existing different curve), hold Ctrl and click on the image. The program also allows selecting multiple types of filament by setting different ClassID. You can interpolate the curve with different spacing by clicking the Interpolate button, but it is only for visualization. After picking, close the small window.
Filament particle extraction
To extract the filament particles, run
e2spt_extract.py --boxsz_unbin=384 --newlabel=fiber0 --threads=8 --alltomograms --shrink=2.0 --tltkeep=1.0 --compressbits=8 --parallel=thread:4 --curves=0 --curves_overlap=.9
Here instead of using --label, we use --curve to specify the type of filament particle we want to extract. The value of --curve is defined by ClassID in the curve drawing interface, and can be viewed in the tomogram evaluation window. --curves_overlap defines the overlap between neighboring boxes along a curve. i.e. --curve_overlap=0 means the boxes do not overlap and 0.5 means each box overlaps with half of the next one. Here we use a large value to increase particle count since we do not have many filaments in the small dataset. Same as other particles, --boxsz_unbin defines the output box size. Since the curve drawing interface does not show boxes, it is a good idea to roughly measure the size of boxes using the regular manual boxer. The newly extracted particles will be labeled by --newlabel.
From this dataset, I found 4 flagella filaments from 3 of the tomograms, and got 263 particles total.
Filament refinement
First, we make the initial model directly from the particles.
e2spt_sgd_new.py sets/fiber0_bin2.lst --res 40 --curve
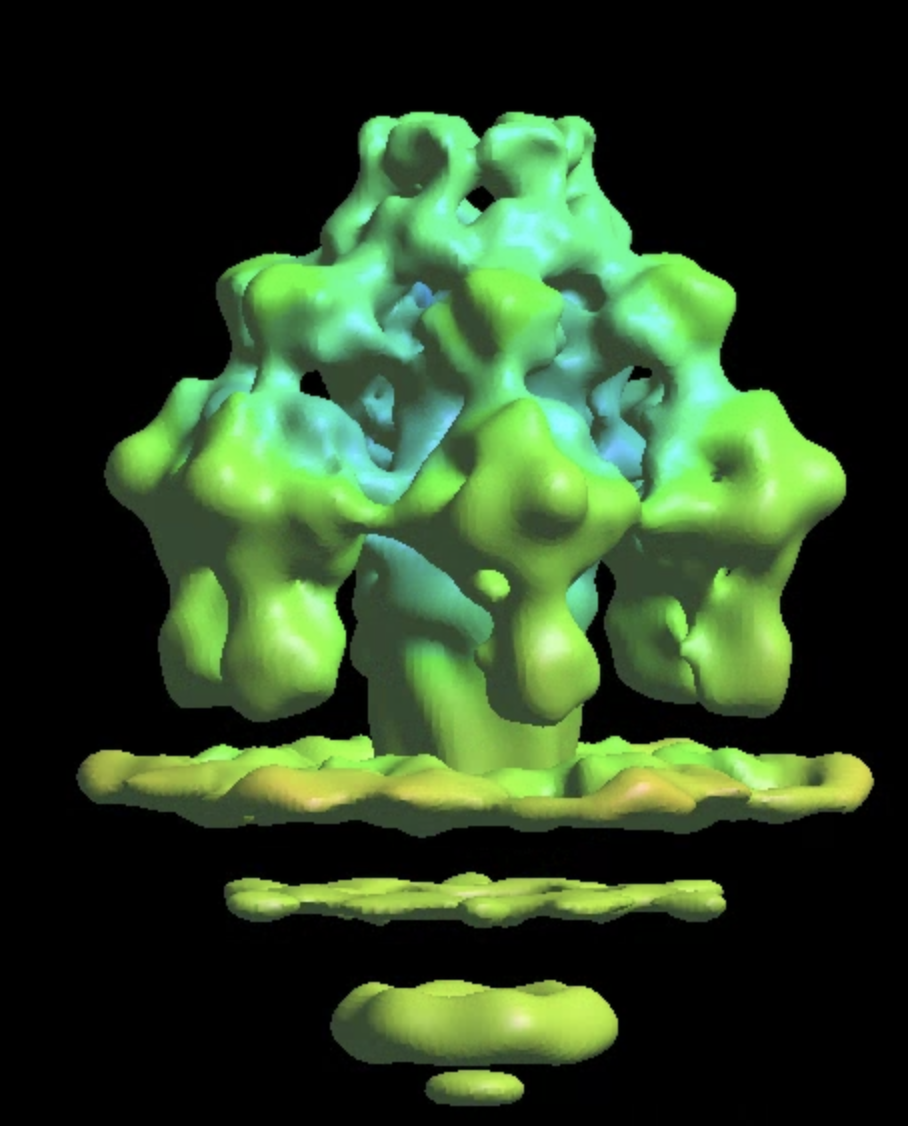
The --curve option will preserve the directional information from the curve and use that for the orientation assignment of particles. This should result in a reasonable initial model with some repeating features.

The flagella have helical symmetry. Normally, we first refine the structure without symmetry, then estimate the symmetry from the subtomogram averaging results, and repeat the refinement with the correct symmetry. However, since we are limited by particle count, it is challenging to get anything with c1. So we simply take the konwn symmetry from existing single particle results from EMDB. Here we use --sym h7:1:65.45:-1.15, where h7 making 7 symmetrical copies, 1 means no additional c symmetry, 65.45 is the helical turn in degrees, and -1.15 is the rise in pixels. Here we add the minus sign before the rise value, since the convention in EMAN2 is different from that used in the EMDB entry. To confirm, download the EMDB map and apply the helical symmetry in EMAN2.
To apply the symmetry, we still need to align the initial model to the symmetry axis. Since we use the information from the curve direction, the structure should already be quite close to the symmetry axis. To get to the correct axis, a simple way is to apply the symmetry first, then align the raw output to the symmetrical one. The process may need to be repeated a few times until it falls into the correct position.
e2proc3d.py sptsgd_01/output_cls0.hdf sptsgd_01/output_cls0a.hdf --sym h7:1:65.45:-1.15 --process mask.soft:outer_radius=-40 e2proc3d.py sptsgd_01/output_cls0.hdf sptsgd_01/output_cls0b.hdf --align rotate_translate_3d_tree --alignref sptsgd_01/output_cls0a.hdf e2proc3d.py sptsgd_01/output_cls0b.hdf sptsgd_01/output_cls0a.hdf --sym h7:1:65.45:-1.15 --process mask.soft:outer_radius=-40
Finally, we can do the refinement using the particles and the initial model.
e2spt_refine_new.py --ptcls sets/fiber0_bin2.lst --ref sptsgd_01/output_cls0_xf_sym.hdf --sym h7:1:65.45:-1.15 --curve --parallel thread:32 --startres 30 --iters=p4,t2 --keep=0.95 --threads=32 --mask mask_cyl.hdf
Similar to the initial model, the --curve option will use the filament curve direction for orientation search. If the filament bends a lot, it is recommended to manually craft a shorter mask (using the mask.cylinder processor) to focus the alignment on a small segment.

